SCREENINGOFNELLORE MAHSURI RICE VARIETYFOR BACTERIALLEAFBLIGHT TOLERANCE USING GENE SPECIFIC SSR MARKERS.
0 Views
P. DIVYA, *N.P. ESWARA REDDY AND A.SRIVIDYA
Department of Molecular Biology and Biotechnology,S.V. Agricultural College, ANGRAU, Tirupati-517502, Chittoor District, Andhra Pradesh, India.
ABSTRACT
Nellore Mahsuri (NLR34449) is a medium fine grain rice variety and mostly adopted by many of the farmers. However, the variety is resistant to blast but susceptible to bacterial leaf blight (BLB). Phenotype screening had been done for two varieties to determine BLB score of Nellore Mahsuri (NLR34449) in comparison to a known check RPBio226 (improved samba Mahsuri), which is having three broad spectrum tolerant BLB genes viz., Xa21, xa13 and xa5. Genotyping of two varieties was studied using three SSR gene specific markers viz., pta248, xa13 pro, xa5FS, linked to the BLB resistance genes viz. Xa21, xa13 and xa5, respectively. Allelic scoring for these markers revealed the abscence of these three broad spectrum genes in NLR34449. Hence, introgression of these genes would improve the tolerance levels of this popular variety.
KEYWORDS:
Bacterial leaf blight (BLB), tolerance, SSR markers
INTRODUCTION
Rice (Oryza sativa L.) is an important food crop
that serves as a major carbohydrate source for nearly half of the world’s population. India is the second largest producer of rice after China with an area of over 43.86 m ha and production of 104.80 mt (Directorate of Economics
& Statistics, 2015). Bacterial leaf blight causes significant yield losses in severely infected fields ranging from 20 to 30 per cent, but this can reach as high as 80 to 100 per cent (Baliyan et al., 2016) and also severely affects the grain quality. Nellore mahsuri is a fine grain rice variety, high yielding, short duration, non-lodging resistant to blast but it is susceptible to BLB. NLR34449 from Agriculture Research Station, Nellore, ANGR Agricultural University.
Chemical control for the management of BLB is
not effective. Therefore, host plant resistance offers the most effective, economical and environmentally safe op-tion for management of BLB (Sombunjitt et al., 2017). Therefore, most researchers are interested in identifying a resistant cultivar and searching for available resistance genes against BLB disease Currently, 40 genes which confer resistance to various Xoo strains have been des-ignated in a series from Xa1 to Xa40 (Sombunjitt et al., 2017 and Kim et al., 2015). A rice cultivar carrying many resistance genes can show a broad spectrum and higher
level of resistance to pathogens than a cultivar with a single resistance gene (Rajpurohit et al., 2010).
MATERIALSAND METHODS
Two rice varieties i.e., NLR34449 (Nellore Mahsuri) and RPBio226 (Improved Samba Mahsuri) were used as parents in this work. Characters of two varieties
listed in the Table 1.
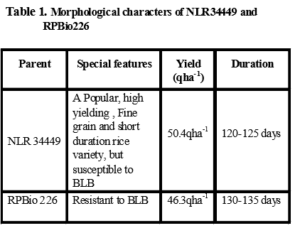
Phenotypic Screening for bacterial leaf blight resistance
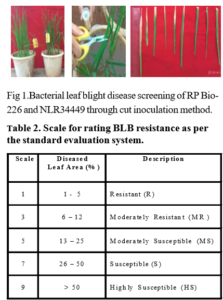
During kharif 2017, parents NLR34449 and RPBio226 were screened against bacterial leaf blight pathogen. The virulant isolate of Xanthomonas oryzae pv. oryzae (Xoo) was collected from Agricultural Re-search Station (ARS), Nellore was used for phenotypical screening of two varieties under glass house conditions. The rice plants were clip-inoculated with a bacterial sus-pension of 109cfu/mL at the maximum tillering stage (50 d after transplanting).Approximately 10 leaves per plant were inoculated, In NLR34449 BLB symptoms were ob-served 2nd day onwards from the day of inoculation. RPBio226 hasn’t shown any symptoms. Nellore Mahsuri was found susceptible for bacterial leaf blight, while im-proved samba mahsuri was resistant, since it carries three resistance genes xa5, xa13 and Xa21 for bacterial leaf blight resistance. Disease severity is assessed based on lesion length measurement or estimation of diseased leaf area. Disease score was recorded based on lesion length as per 1-9 scale of standard standard evaluation system
(SES) score (IRRI, 2013) as shown in Table 2.
Genotyping
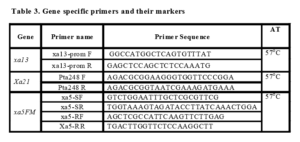
Genomic DNA was extracted from freshly collected leaves of the two parents, NLR34449 and RPBio226 by using CTAB method. The PCR amplification using SSR primers was carried out using sterilized thin walled PCR tubes (0.2 ml). PCR protocol for amplification of markers (pTA248, xa13 pro, xa5FS) as described by Sundaram et al. (2008) was adopted. As regards to the newly designed functional markers for xa13 and xa5, PCR was performed using 0.1 µl of(5U/µl) Taq DNA polymerase (abm),2 µL of (100ng/ µL) DNA template DNA, 0.5 µL OF 5mM of each primer, µl of 1Mm dNTPs, and 1 µl 10X PCR buffer (abm) in 25 µL reaction volume with a thermal profile of 94 °C for 5 min (initial denaturation), followed by 30 cycles of denaturation at 94 °C for 30 seconds, annealing at 55 °C for 30 seconds, extension at 72 °C for 1 min and a final extension of 7 min at 72 °C. The amplified products were electrophoretically resolved on a 2.0 per cent agarose gel, stained with ethidium bromide and visualized in a gel documentation system (Alpha Innotech, USA). However, with respect to the multiplex PCR assay for simultaneous detection of Xa21, xa13 and xa5, the protocol was essentially same as described above, except that annealing was done at 57 °C and the amplified products were resolved in 2 per cent agarose gels. The primer sequences for the functional markers for the three resistance genes are given in Table 3
RESULTS AND DISCUSSION
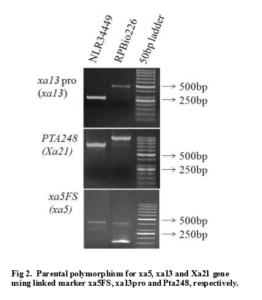
The DNAs from the recurrent parent NLR34449 and the donor parent RPBio226 (for xa21, xa13 and xa5) were used to determine marker polymorphism. The reported sequenced tagged sites (STS) markers xa5FS, xa13pro and Pta248 showed polymorphism between recurrent parent Nellore Mahsuri and donor parent RPBio226. The primer pair pta248 amplified fragments of 950bp in the resistant parent RPBio226 (improved samba mahsuri), while that from the susceptible parent NLR344449 was 660bp. With respect to the primer pair, xa13-prom, RPBio226 (improved samba mahsuri), amplified a 500bp fragment, while NLR34449 amplified a 250bp. Similarly, with respect to the marker xa5FS, a fragment of 313bp was amplified in NLR34449, while a 420bp and 150bp fragments were amplified in RPBio226. Parental polymorphism between NLR34449 and RPBio226 (improved samba mashuri) was observed with the xa4, xa5, xa8, xa13, xa21, xa23 gene specific markers (Fig. 2) of which, all the genes, except xa8,showed polymorphism between the parents.
CONCLUSION
Based on allelic pattern of BLB resistant genes NLR34449 (Nellore mahsuri) rice variety is susceptible to BLB. Major BlB genes are transferred into NLR34449 through Marker assisted introgression and developed into a resistant variety.
LITERATURE CITED
- Baliyan, N., Mehta, K., Rani, R., Purushottum, Boora, K.S. 2016. Evaluation of Pyramided Rice Genotypes Derived from Cross Between CSR30 and IRBB60 Basmati Variety against Bacterial Leaf Blight. Vegetos. 29: 3.
- Kim, S., Suh, J., Qin, Y., Noh, T., Reinke, R.F and Jena, K.K. 2015. Identification and fine24 mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races 25 in rice (Oryza Sativa L.). Theoretical and Applied Genetics. 128: 1933–1943.
Rajpurohit, D., Kumar, R., Kumar, M., Paul, P., Awasthi, A., Basha, P.O., Puri, A., Jhang, T., Singh, K. and Dhaliwal, H.S. 2010. Pyramiding of two bacterial blight resistance and a semidwarfing gene in Type 3 Basmati using markerassisted selection. Euphytica. 178(1):111-126. - Sombunjitt, S., Sriwongchai, T., Kuleung, C., Hongtrakul, V .2017. Searching for and analysis of bacterial blight resistance genes from Thailand rice germplasm. Agriculture and Natural Resources. 51 : 365-375
Sundaram R. M., Vishnupriya M. R., Biradar S. K., Laha G. S., Reddy A. G., Rani N. S., Sarma, N.P and Sonti, R.V. 2008 Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica. 160: 411– 422.
- Bio-Formulations for Plant Growth-Promoting Streptomyces SP.
- Brand Preference of Farmers for Maize Seed
- Issues That Consumer Experience Towards Online Food Delivery (Ofd) Services in Tirupati City
- Influence of High Density Planting on Yield Parameters of Super Early and Mid Early Varieties of Redgram (Cajanus Cajan (L.) Millsp.)
- Influence of Iron, Zinc and Supplemental N P K on Yield and Yield Attributes of Dry Direct Sown Rice
- Effect of Soil and Foliar Application of Nutrients on the Performance of Bold Seeded Groundnut (Arachis Hypogaea L.)

