Prevalence of Sugarcane Yellow Leaf Virus In Commercially Grown Sugarcane Varieties in Andhra Pradesh
0 Views
T.M. HEMALATHA*, B.V. BHASKARA REDDY, R. SARADA JAYALAKSHMI,
S.R. KOTESWARA RAO, M. HEMANTH KUMAR AND G. MOHAN NAIDU
Agricultural Research Station, ANGRAU, Perumallapallae – 517 505
ABSTRACT
Sugarcane Yellow Leaf Disease caused by Sugarcane Yellow Leaf Virus (SCYLV) is a serious viral disease affecting production and productivity in many ruling sugarcane varieties in Andhra Pradesh. SCYLV is a single stranded, positive sense, RNA genome, with six open reading frames (ORFs 0-5) and three un-translated regions (UTRs) consisting of 5.8 kb nucleotides. The losses due to the disease range from 60 per cent in the main crop to 100 per cent in the ratoon crop. Twenty five SCYLV infected leaf samples were collected from major sugarcane growing regions and amplified the ORF 1& 2 regions of the genome of the virus coding for RNA dependent RNA polymerase gene (RdRp) using the primer pair (SCYLV-F3 and SCYLV-R3). Positive amplifications of 1110 bp obtained from twelve samples were sequenced and analyzed for sequence identities, sequence variations and phylogenetic relationships with the SCYLV isolates reported earlier from India and other countries with the view to determine the variability among the SCYLV isolates reported from India and abroad. The nucleotide (nt) and amino acid (aa) sequence comparison in the RdRp coding regions showed a significant variation between Indian isolates (SCYLVIND) and other SCYLV isolates reported worldwide. The multiple sequence analysis of the isolates under the study with 22 similar sequences of SYLCV isolates reported from India and worldwide revealed 90.6 – 100% identities with IND, REU, BRA, CHN, HAW, CUB, COL and PER isolates reported from other countries. The nucleotide identities were 95.7 – 100 per cent with other SCYLV isolates reported from India. There was greater similarity in amino acid sequences i.e., up to 94.2 – 99.2 per cent between the Indian isolates and the CUB, COL isolates while the similarity was about 90.0 – 96.1 per cent with other genotypes. This was further confirmed from the phylogenetic analysis where the CUB and COL isolates clustered with the isolates reported from India while the other genotypes (HAW, CHN, REU, PER, BRA) clustered into a separate group. The Indian isolates showed close relationship with the CUB and COL isolates. Hence it was confirmed from the study that SCYLV isolates collected from major sugarcane growing regions of Andhra Pradesh are closely related to SCYLV-CUB and SCYLV-COL isolates from Cuba and Columbia respectively.
KEYWORDS:
Sugarcane, Sugarcane yellow leaf virus, Molecular Characterization, Movement, Protein, Phylogenetic analysis.
INTRODUCTION
Sugarcane yellow leaf virus (SCYLV) is a distinct member of the Polerovirus genus of the Luteoviridae family. SCYLV is the major limitation to sugarcane production worldwide and presently posing a major threat to sugarcane cultivation. The disease was first reported in Hawaii in the late 1988 and was subsequently observed in almost all sugarcane growing countries (Comstock et al., 2002; Vega et al., 1997; Viswanathan, 2002). In India, it was reported during 1999 (Viswanathan, 2002). The disease was reported from almost all the sugarcane growing regions in India (Maharashtra, Bihar, Uttar Pradesh, Punjab, Kerala, Tamil Nadu, Madhya Pradesh, Haryana and Andhra Pradesh). Disease incidence up to 100 per cent in commercial fields in susceptible cultivars was reported in Florida (Comstock et al., 1999 and 2001), India (Viswanathan, 2002), Island of Reunion (Rassaby et al. 2004) and Thailand (Lehrer et al. 2008).
SCYLV is an emerging virus that has evolved by recombination between ancestors of the three genera (Luteovirus, Polerovirus, and Enamovirus) forming the family Luteoviridae. The genome of SCYLV has a positive- sense single stranded RNA containing six overlapping open reading frames (ORF 0– ORF 5) which is devoid of a poly(A) tail and three untranslated regions (UTRs) consisting of ~5.8 kb nucleotides. P0 (ORF0) codes for a suppressor of host RNA silencing (Abu Ahmad et al., 2006). ORF1 overlaps ORF0 and ORF2 in the 5′ and 3′ termini, respectively. An ORF1/ORF2 is a fusion protein translated via a frame shift, producing the RNA dependent RNA polymerase (RdRp). ORF4, which encodes the movement protein, is located within ORF3 which codes for coat protein. ORF5 is expressed as read through protein with ORF3 and it codes for an aphid transmission factor.
In Andhra Pradesh, sugarcane is grown in an area of 1.26 lakh hectares with average production of 76.14 t ha-1 (Source. Cooperative Sugar vol. 51 issue 6, Feb, 2020). The disease was first recorded in Nizamabad area during the year 2004 (Bharathi and Kishan Reddy, 2007). The wide prevalence of the disease was observed during 200809 season in various sugarcane varieties used in All India Coordinated trials at Agricultural Research Station, Perumallapalle, Tirupati, (Andhra Pradesh). Soon, the disease was noticed on other commercially grown varieties viz., 2003 V46, 86 V 96, Co 7219, 87 A 298, Co 86032 in Coastal and Southern regions of Andhra Pradesh. At present the disease is spreading at an alarming rate infecting almost all the varieties grown by the farmers in A.P. The disease has spread to number of ruling varieties like- 2003V46, Co 86032, 83V15, 87A298, 86V96, Co 62175, 2002V48, 2005T16. The percent disease incidence up to 30.6 per cent in plant crop and 51.0 per cent in ratoon crops was recorded in most of the cultivated varieties (Hemalatha et al., 2014; Suresh et al., 2020).
Studies regarding phylogenetic origin of SCYLV revealed that 10 different genetic groups have determined viz., BRA (Brazil), CHN1, CHN2, CHN3 (China), COL (Colombia), CUB (Cuba), HAW (Hawaii), IND (India), PER (Peru) and REU (Reunion). These genotypes were determined based on analysis of the genetic diversity of their genome using partial sequences and complete genomes (Moonan and Mirkov, 2002; Abu Ahmed et al., 2006a; Elsayed et al., 2010; Gao et al., 2012, Chinnaraja et al, 2013; Lin et al. 2014).
The main objective of the present study was to detect the prevalence and distribution of Sugarcane yellow leaf virus in sugarcane growing regions of Andhra Pradesh and to determine the diversity of SCYLV isolates in India and their phylogenetic origin.
MATERIAL AND METHODS
During 2017, twenty five sugarcane leaf samples exhibiting typical midrib yellowing symptoms were collected from major sugarcane growing regions of Andhra Pradesh (Chittoor, Nellore, Vishakapatnam, Krishna, Prakasham and Vijayanagaram). The collected leaf samples were stored at -20°C until further processing. The details of the SCYLV isolates used in the study are given in Table 1.
RNA extraction and amplification of ORF1 & 2 regions of the virus genome from the infected samples.
One hundred mg of fresh leaf sample powdered using liquid nitrogen was transferred to a 1.5 ml DEPC treated microfuge tube and to that 1 ml TRI reagent (Sigma, USA) was added. The tubes were vigorously shaken for homogenous mixing of TRI reagent with the sample and were kept at 4°C until all the samples were homogenized. The samples were allowed to complete dissociation of nucleoprotein complexes and release of RNA by incubating at room temperature for 5 min. All the insoluble materials such as cellular membranes, high molecular weight DNA and polysaccharides were precipitated at the bottom of the tubes by centrifugation at 12,000 rpm for 10 min at 4°C. The supernatant containing the RNA was transferred to a fresh tube and to that 200 ìl of the chloroform for every 1 ml of TRI reagent was added. The tubes were shaken vigorously for 15 sec and again incubated at room temperature for 5–10 min and later centrifuged at 12,000 rpm for 15 min at 4°C. After centrifugation, the mixture separated into three phases: a red organic phenolic phase contained protein, an interphase of DNA and a colorless upper aqueous phase which contained the RNA. The RNA

containing upper aqueous phase was transferred into a fresh tube and added 500 ìl of isopropanol to precipitate the RNA. After incubation for 5 min at room temperature, the tubes were centrifuged at 12,000 rpm for 15 min at 4°C for pelletizing the RNA.
The pellet was air dried for 10 min and dissolved the RNA with 40 ìl of RNA resuspension buffer (Ambion, USA) and stored at “80°C.
The quality of the RNA was checked by
Nanospectrophotometer. The forward primer SCYLV- F3:
52 -GCAGCAGAACGGAGGGAAGAAGTC3′ and reverse primer SCYLV-R3:5′-TGAGTTTGGGC GTACARGACACCGCC3′ designed by Viswanathan et al. (2008) were used to amplify ~1110 bp of ORF1 and ORF2 regions of SCYLV genome. The cDNA was synthesized from total RNA of all the 25 samples separately using Revert Aid H Minus first stand cDNA synthesis kit (MBI Fermentas, USA) primed with 50 pmol of SCYLV-R3 by following the manufacturer’s protocol. The PCR reaction was performed in a total volume of 25 ìl containing 2 ìl cDNA, 2.5 ìl of 10×PCR buffer containing 15 mM MgCl2, 0.5 ìl of 10 mM dNTP mix, 10 pmol each of forward and reverse primers (SCYLV-F3 & SCYLV- R3), 1.25 units of Taq polymerase (Bangalore Genei, Bangalore), and sterile miliQ water to a final volume. 30 PCR cycles were performed in a PCR thermocycler (Mastercycler gradient; Eppendorf, Germany) with the conditions: initial denaturation at 94°C for 4 min, denaturation at 94°C for 1 min, primer annealing at 65°C for 1 min, extension at 72°C for 45 sec with a final 72°C extension for 10 min. A 10 ìl aliquot of each amplified product was analyzed by electrophoresis on 1.5 per cent agarose gels stained with eithidium bromide.
Sequence analysis and phylogenetic relationship
cDNA from 12 positive samples were amplified with SCYLV-F3 and SCYLV-R3 by RT-PCR. The amplicon of 1110 bp from 12 samples were eluted using GenElute Gel Extraction Kit (Sigma, USA). The nucleotide sequence of each virus isolate was sequenced with virus specific primers for ORF1 and ORF2 regions. The nucleotide sequence data of 12 SCYLV isolates were analyzed using Bioedit and MEGA 7.0 (Kumar et al., 2016) to study the sequence identities/similarities with the other 22 SCYLV isolates available in GenBank database (Table 2) including partial sequence data of RdRp of representative genotypes (BRA from Brazil, CUB from Cuba, PER from Peru, and REU from Reunion Island). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) were shown next to the branches and phylogenetic tree was generated with Neighbour-Joining method.
RESULTS AND DISCUSSION
The typical symptoms of the disease observed were intense yellowing of midribs on the abaxial surface, lateral spread of yellow discoloration to the leaf lamina followed by tissue necrosis from the leaf tip spreading downwards along the midrib and a bushy appearance of the top of the plant due to internode shortening in maturing plants (Fig. 1a and 1b). In some sugarcane cultivars, leaves show a pinkish discoloration of the midrib on the adaxial surface.


Figure 1A & B. A. Typical symptoms of midrib yellowing in Sugarcane plant infected with Sugarcane yellow leaf disease. B. Inter nodal shortening of the plant.
The disease incidence was significantly high (60–70%) at later stage (6–7 months of age) of the crop. Similar symptoms of sugarcane yellow leaf disease was observed by Bharathi and Reddy (2007) in sugarcane varieties Co 6907 and Co C 671 in Northern Telangana zone of Andhra Pradesh. The symptoms were evident in mature leaves especially during the month of September. The incidence of disease varied from 3 to 36 per cent in different cultivars with maximum incidence in Co 6907 and Co C 671. Singh et al., (2018) surveyed for Yellow leaf disease in sugarcane fields of Uttar Pradesh and revealed that the incidence of the disease was up to 30% and mostly appeared during 68 months age of the crop.
Detection of SCYLV by RT-PCR
The expected size (1110 bp) fragment was successfully amplified with SCYLV-F3 and SCYLV-R3 primers by RT-PCR in 12 symptomatic samples collected from major sugarcane growing regions of Andhra Pradesh i.e. CO 86032, 93A145, 2003V46, 83V15, 87A298, 2005T16, 2009A240, 2009V89, CO T 8201, 92A30, CoC 671 (Fig. 2).
Successful PCR detection of RdRp gene indicated that the SCYLV was wide spread among the most popular varieties grown in major sugarcane growing regions throughout India. Similar reports are available on the widespread occurrence of SCYLV in other parts of the world (Arocha et al., 2005; Fitch et al., 2001; Rott et al., 2008).
Sequencing and Phylogenetic analysis
Amplicons of 1110 bp with prominent intensity obtained from infected leaf samples were sequenced at Eurofins India, Banglore. The BLAST analysis of partial RdRp gene sequences obtained from 12 different cultivars showing >90 per cent sequence identity with SCYLV genomes from NCBI data base were deposited in
GenBank database. The sequence data of 12 isolates under study was deposited in GenBank database under accession numbers: MN865128 (SCYLV-SKHT1-2003V46), MN909725 (SCYLV-YPD1-83V15), MN900951 (SCYLV-PTR-2005T16), MT060294 (SCYLV-YPDCoT8201)), MN972615 (SCYLV-SKHT2-86V96),
MT074088 (SCYLV-ANK1-87A298) , MN857470 (SCYLV-ANK2-93A145), MT909726 (SCYLV-PPL1-
92A30), MN865129 (SCYLV-PPL2-Co 86032), MN865130 (SCYLV-PPL3-CoC 671), MW031686 (SCYLV-VYV-2009V89), MT992722 (SCYLV-ANK32000A240).
The multiple sequence analysis of the isolates under the study with 22 similar sequences of SYLCV isolates reported from India and abroad revealed 90.6-100 per cent
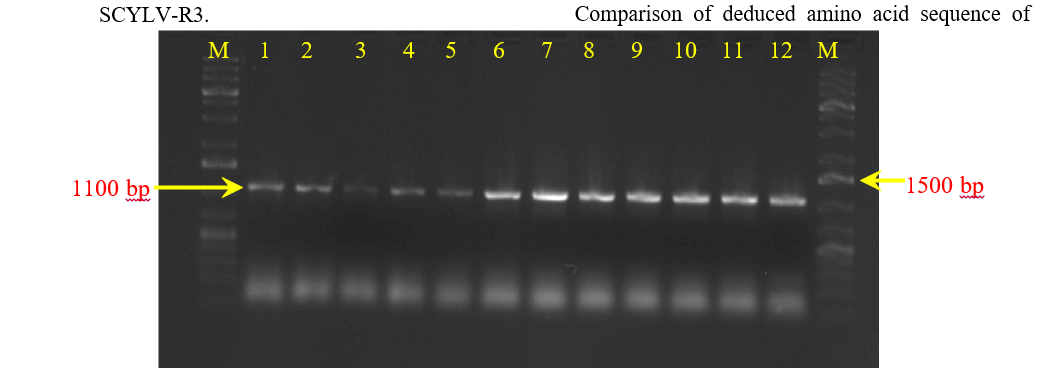
Figure 2. Amplification of ORF1&2 regions of 12 isolates of SCYLV using specific primers SCYLV-F3 and
identities with IND, REU, BRA, CHN, HAW, CUB, COL and PER genotypes reported from other countries. The nucleotides (nt) and their deduced amino acid (aa) sequences of ORF 1 & 2 of the isolates under the study were compared with similar sequences of SCYLV isolates available in GenBank database. The isolates under the study shared 95.7–100% nucleotide identities among them and with other isolates SCYLV reported from India (Table. 3). The highest nucleotide similarity of 100% was observed between SCYLV-YPD2- Co T 8201 (MT060294) and SCYLV-VYV-2009V89 (MW032686) isolates of Andhra Pradesh and between IND-LCKNW-
KF680098 and SCYLV-SKHT1-2003V46 (MN865128); IND-TN-EU624496 and SCYLV-ANK1-87A298 (MT074088); IND- EU624497 and SCYLV-PPL2-Co 86032 (MN865129); JF925155 and SCYLV-YPD183V15 (MN909725) isolates of India. All the Indian isolates shared maximum identity with SCYLV-CUB isolate (MF622079) and SCYLV-COL isolate
(MF622078) with 96.6-99.0 % while they shared 90.6– 92.2 per cent nucleotide identity with SCYLV isolates (REU, BRA and PER genotypes) from other parts of the world.
The nucleotide sequence data presented here are in consistent with previous reports which include SCYLV, the causative agent of the yellow leaf in Brazil, USA, Australia, and Mauritius as a possible member Polerovirus of the family Luteoviridae (Rott et al., 2008; Scagliusi and Lockhart, 2000). The sequence comparisons reported in this study contribute to a better understanding of the taxonomic status of SCYLV isolates throughout the world.
Comparison of deduced amino acid sequence of RdRp among the 12 SCYLV isolates under the study revealed maximum similarity ranging from 93.40 – 100.00 per cent with IND isolates while 93.40 – 98.30 per cent similarity with SCYLV-CUB genotype (MF622079) and 93.00 – 98.30 per cent with SCYLV-COL genotype (MF622078).The other genotypes viz. CHN, REU, BRA, PER, HAW shared 86.10 – 90.60 per cent amino acid similarity with the IND isolates under the study (Table 4).The SCYLV isolates from Cuba and Colombia showed closest homology with the Indian isolates. These findings suggest that the mixed populations of SCYLV isolates that exist across India, may be due to the movement of the SCYLV isolates within the country through infected propagative material. In a study that included 18 geographical locations worldwide, the BRA-PER strain occurred in most sugarcane producing areas wherever as genotypes CUB and REU were found in four geographical locations only. Afterwards, several isolates of SCYLV were detected in Brazil, Colombia, Guadeloupe, Mauritius and Reunion Island, suggesting different virus introductions and/or different evolution histories of the virus after its introduction into a new environment (Abu Ahmad et al. 2006 b). On the basis of SCYLV genotypes identified in Brazil, Colombia, Cuba and Peru, Abu Ahmad et al.(2006 b) suggested that they may have been introduced through infected planting material imported from elsewhere.
Phylogenetic analysis revealed that the SCYLV isolates under the study clustered into two major groups. SCYLV isolates from India, Colombia and Cuba clustered

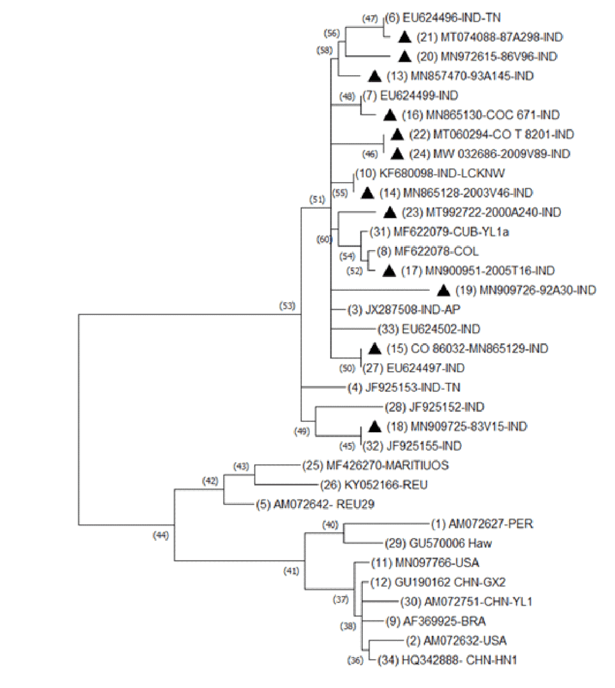
Figure 3. Phylogenetic tree constructed using maximum likelihood method of MEGA version 7.0. based on the RdRp gene sequences of SCYLV isolates at nucleotide level.
into one group and all other isolates from the other countries i.e., China, Brazil, Peru, Reunion, Kenya, Hawaii, USA formed into a separate cluster (Fig. 3). The 12 SCYLV isolates under study showed close relationship with Cuba isolate (MF622079) and Colombia
(MF622078) isolate along with other nine isolates from India. The 13 SCYLV isolates reported from other parts of the world formed into a separate cluster with independent clades: HAW, USA, MARITIUS, REU, KENYA, CHN, BRA, REU and PER.
Viswanathan et al. (2008) studied the phylogentic analysis of SCYLV isolates based on partial ORF 1 and 2 sequences of the virus genome. The sequence analysis suggested that the population that exists in India was significantly different from rest of the world. It was revealed from his study that CUB genotype was predominant among four genotypes (BRA, CUB, IND and PER) found in India.
It is remarkable to note that the isolates of India clustered with isolates of Cuba and Colombia and shared maximum sequence similarity upto 99.2 per cent . Similarly, Moonan and Mirkov (2002) revealed from their study that SCYLV was widespread in most of the sugarcane producing countries. The virus isolates collected from North, South and Central America were divided into two groups: one group contained only isolates from Colombia (C-population) and the other group contained the isolates from Argentina, Brazil, Guatemala, USA, Florida, Louisiana and Texas.
The phylogenetic analyses of the entire genome of SCYLV described by Abu Ahmad et al. (2006a) revealed the occurrence of three genotypes of SCYLV (BRA, PER and REU) based on the geographical location where it was first detected; Brazil, Peru and Reunion, respectively. Additionally, a virus isolate from Cuba (that was partially sequenced) showed only 77.00 – 80.00 per cent amino acid sequence identities in ORF1 with isolates of genotypes BRA, PER and REU, which suggest that the Cuban isolate represent a new genotype (CUB).
It is evident from the study that Sugarcane yellow leaf disease is threatening the sugarcane cultivation affecting almost all the varieties grown in India and abroad. The phylogenetic analysis revealed that the SCYLV isolates reported from India shared maximum nucleotide and amino acid similarity with the SCYLVCuban and Colombia isolates. The diversity among the SCYLV isolates used in the study showed a very less variation between them while the variability was greater with the BRA, CHN, REU, PER, HAW isolates.
LITERATURE CITED
Abu Ahmad, Y., Rassably, L., Royer, M., Borg, Z., Braithwaite, K.S and Mirkov, T. 2006a. Yellow leaf of sugarcane is caused by at least three different genotypes of Sugarcane yellow leaf virus, one of which predominates on the Island of Réunion. Archives of Virology. 151: 1355-1371.
Abu Ahmad, Y., Royer, M., Daugrois, J.H., Costet, L., Lett, J.M., Victoria, J.I., Girard, J.C and Rott, P. 2006b. Geographical distribution of four Sugarcane yellow leaf virus genotypes. Plant Disease. 90: 11561160.
Arocha, Y., Lopez, M., Fernandez, M., Pinol, B., Horta,
D., Peralta, E.L., Almeida, R., Carvajal, O., Picornell, S., Wilson, M.R and Jones, P. 2005. Transmission of a sugarcane yellow leaf phytoplasma by the delphacid plant hopper Saccharosydne saccharivora, a new vector of sugarcane yellow leaf syndrome. Plant Pathology. 54: 634-642.
Bharathi, V and Kishan Reddy, L. 2007. Mixed infection of sugarcane streak mosaic and sugarcane yellow leaf virus infecting sugarcane crops in Andhra Pradesh, India. International Journal of Agricultural Sciences 3(1): 68-72.
Chinnaraja, C., Viswanathan, R., Karuppaiah, R., Bagyalakshmi, K., Malathi, P and Parameswari, B. 2013. Complete genome characterization of sugarcane yellow leaf virus from India: Evidence for RNA recombination. European Journal of Plant Pathology. 135: 335-349.
Comstock, J.C., Miller, J.D and Schnell, R.J. 2001. Incidence of Sugarcane yellow leaf virus in clones maintained in the world collection of sugarcane and related grasses at the United States National Repository in Miami, Florida. Sugar Tech. 3: 128133.
Comstock, J.C., Miller, J.D., Tai, P.Y.P and Follis, E.J. 1999. Incidence of and resistance to Sugarcane yellow leaf virus in Florida. Proceedings of International Society of Sugar Cane Technology. 23: 366-372.
Comstock, J.C., Pena, M.J., Vega, A.F and Lockhart, B.E.L. 2002. Report of sugarcane yellow leaf virus (SCYLV) in Ecuador, Guatemala and Nicaragua. Plant Disease. 86:74.
El Sayed, A.I., Weig, A.R and Komor, E. 2010. Molecular characterization of Hawaiian Sugarcane yellow virus leaf genotypes and their phylogenetic relationship to strains from other sugarcane growing countries.
European Journal of Plant Pathology. 129:399-412.
Fitch, M.M.M., Lehrer, A.T., Komor, E and Moore, P.H. 2001. Elimination of Sugarcane yellow leaf virus from infected sugarcane plants by meristem tip culture visualized by tissue blot immunoassay. Plant Pathology. 50: 676-680.
Gao, S.J., Lin, Y.H., Pan, Y.B., Damaj, M.B., Wang, Q.N., Mirkov, T.E and Chen, R.K. 2012. Molecular characterization and phylogenetic analysis of Sugarcane yellow leaf virus isolates from China. Virus Genes. 45: 340-349.
Hemalatha, T.M., Kumar, M.H.I., Afsar, S., Reddy, B.V.B and Rao, M.S. 2014. Molecular detection of Sugarcane Yellow Leaf Virus in popularly grown sugarcane varieties in Andhra Pradesh and characterization of the virus from the Sugarcane variety 2003V46. Journal of Mycology and Plant Pathology. 44(4): 442-446.
Kumar, S., Stecher, G and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 33(7): 1870-1874.
Lehrer, A.T., Kusalwong, A and Komor, E. 2008. High incidence of Sugarcane Yellow Leaf Virus (SCYLV) in sugar plantations and germplasm collections in Thailand. Australasian Plant Disease Notes. 3: 8992.
Lin, Y.H., Gao, S.J., Damaj, M.B., Fu, H.Y., Chen, R.K and Mirkov, T.E. 2014. Genome characterization of Sugarcane yellow leaf virus from China reveals a novel recombinant genotype. Archives of Virology .159: 1421-1429.
Moonan, F and Mirkov, T.E. 2002. Analyses of genotypic diversity among North, South, and central american isolates of Sugarcane yellow leaf virus: evidence for
Colombian origins and for intraspecific spatial phylogenetic variation. Journal of Virology. 76:
1339-1348.
Rassaby, L., Girard, J.C., Lemaire, O., Costet, L., Irey,
M.S., Kodja, H., Lockart, B.E.L and Rott, P. 2004. Spread of Sugarcane yellow leaf virus in sugarcane plants and fields on the island of Reunion. Plant Pathology. 53:117-125.
Rott, P., Mirkov, T.E., Schenck, S and Girard, J.C. 2008. Recent advances in research on Sugarcane yellow leaf virus, the causal agent of sugarcane yellow leaf.
Sugar Cane International. 26:18–27.
Scagliusi, S.M.M and Lockhart, B.E.L. 2000.
Transmission, characterization, and serology of a luteovirus associated with yellow leaf syndrome of sugarcane. Phytopathology. 90: 120-124.
Singh, S.P., Vishwakarma, S.K and Singh, A. 2018. Deterioration in Qualitative and Quantitative Parameters of Sugarcane due to Yellow Leaf Disease. International Journal of Current Microbiology and Applied Sciences. 7(12): 2320-2326.
Suresh, M., Umadevi, G., Kumar, N.R and Tirupathi, M. 2020. Current Status of Yellow Leaf Disease (YLD) of Sugarcane Caused by Sugarcane Yellow Leaf Virus (ScYLV) in the states of Andhra Pradesh and Telangana. International Journal of Bio-resource and
Stress Management. 11(1):073-081
Vega, J., Scagluisi, S.M.M and Ulian, E.C. 1997. Sugarcane yellow leaf disease in Brazil: Evidence of association with a luteovirus. Plant Disease. 81:21-26.
Viswanathan, R., Balamuralikrishnan, M and Karuppaiah, R. 2008. Identification of three genotypes of Sugarcane yellow leaf virus causing yellow leaf disease from India and their molecular characterization. Virus Genes. 37: 368-379.
Viswanathan, R. 2002. Sugarcane yellow leaf syndrome in India: Incidence and effect on yield parameters.
Sugar Cane International. 17-23.
- Bio-Formulations for Plant Growth-Promoting Streptomyces SP.
- Brand Preference of Farmers for Maize Seed
- Issues That Consumer Experience Towards Online Food Delivery (Ofd) Services in Tirupati City
- Influence of High Density Planting on Yield Parameters of Super Early and Mid Early Varieties of Redgram (Cajanus Cajan (L.) Millsp.)
- Influence of Iron, Zinc and Supplemental N P K on Yield and Yield Attributes of Dry Direct Sown Rice
- Effect of Soil and Foliar Application of Nutrients on the Performance of Bold Seeded Groundnut (Arachis Hypogaea L.)

