VARIABILITY, CHARACTER ASSOCIATION AND PATH ANALYSIS STUDIES IN LITTLE MILLET (Panicum sumatrense)
0 Views
L. MADHAVILATHA*, M. SUBBA RAO, M. SHANTHI PRIYA AND M. HEMANTH KUMAR
Department of Genetics and Plant Breeding, Agricultural Research Station, Perumallapalle, Tirupati – 517 505.
ABSTRACT
An experiment was carried out with fifty little millet elite germplasm lines to estimate genetic parameters, character association and path analysis for nine quantitative traits. High GCV and PCV were recorded for grain yield per plant and number of effective tillers per plant. High heritability coupled with high genetic advance was observed for grain yield per plant and number of effective tillers per plant indicating the importance of additive gene action in governing the inheritance of these traits. Character association studies revealed that plant height, length of inflorescence, number of effective tillers per plant and days to 50% flowering showed high positive and significant correlation with grain yield per plant at both phenotypic and genotypic levels. Path analysis revealed that plant height, number of effective tillers per plant and length of inflorescence showed positive association and direct effect on grain yield per plant at both phenotypic and genotypic levels.
KEYWORDS:
Little millet, genetic parameters, correlation and path analysis
INTRODUCTION
Little millet (Panicum sumatrense), belongs to the family Poaceae, sub-family Panicoideae and the tribe Paniceae. It is native to India and is also called Indian millet. The species name is based on a specimen collected from Sumatra (Indonesia). In India, Little millet is one of the important minor cereal crop mainly grown in Karnataka, Tamil Nadu, Orissa, Madhya Pradesh and Andhra Pradesh. It is presently grown throughout India in more than half a million hectares (Nirmalakumari et al., 2010). Little millet is adapted to both temperate and tropical climates. It can withstand both drought and water logging. It is an important catch crop in some tribal farms in India. The production of this crop is low mainly due to the non-availability of improved varieties and nonadoption of production strategies (Suryanarayana and Sekhar, 2018). Wider range of genetic variability helps in selecting desired genotypes. Genetic variability together with the heritability estimates would give a better idea on the amount of genetic gain expected out of selection (Burton, 1952). Heritability estimates along with genetic advance are more helpful in predicting the gain under selection than heritability estimates alone. However, it is not necessary that a character showing high heritability will always exhibit high genetic advance (Johnson et al., 1955). Grain yield is a complex character and is the end product of various traits. Therefore knowledge regarding the correlation of grain yield with other component characters is valuable for understanding the correlated response to selection for yield. Correlation coefficients measures the mutual relationship between various plant characters and determines the component characters on which selection can be based for genetic improvement in yield. Path coefficient analysis is helpful to recognize direct and indirect causes of correlation and also enables us to compare the causal factors on the genetic basis of their relative contributions. Direct effects are where a trait directly affects another without being influenced by other traits whereas indirect effects occur when the relationship between two traits is mediated by one or more traits. Knowledge of the associations between yield and its component traits and among the component traits themselves would allow for more effective selection for yield. Hence the present study of variability, correlation and path analysis would serve path for future breeding programme.
MATERIAL AND METHODS
An experiment was conducted with fifty little millet elite germpalsm accessions at Agricultural Research Station, Perumallapalle during kharif, 2014. The design adopted was RCBD with two replications. Each entry was grown in two rows of 3 m length by adopting a spacing of 22.5 × 10 cm. Recommended package was practiced to raise healthy crop. Observations were recorded on five randomly selected plants for nine quantitative traits viz.,days to 50% flowering, plant height (cm),number of effective tillers per plant, flag leaf length (cm), flag leaf width (cm), length of peduncle (cm), length of inflorescence (cm), length of lower raceme (cm) and grain yield per plant (gm). The data were subjected to statistical analysis for estimation of genetic parameters. Phenotypic and genotypic coefficients of variation (PCV and GCV) were computed according to Burton and DeVane (1953). Heritability in broad sense was estimated as per Allard (1960). Genetic advance was estimated as per the formula proposed by Lush (1940) and genetic advance was expressed as per cent of mean by using the formula suggested by Johnson et al. (1955). Correlation coefficients were worked out using the formula as suggested by Falconer (1981). Further correlation coefficients were further partitioned into direct and indirect effects with the help of path coefficient analysis as suggested by Dewey and Lu (1959).
RESULTS AND DISCUSSION
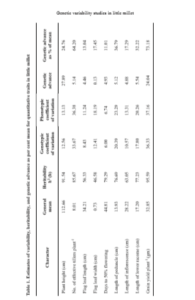
Analysis of variance revealed the significant differences among the genotypes indicating the existence of sufficient variability for all the nine traits studied. Similar substantial variability was reported by earlier workers (Sasamala et al., 2015; Selvi et al., 2015). The estimates on genotypic and phenotypic coefficients of variation, heritability and genetic advance as per cent of mean are furnished in Table 1. The genotypic coefficients of variation were lower than phenotypic coefficients of variation for all the characters indicating the interaction of genotypes with environment. However narrow difference between PCV and GCV indicated little influence of environment on the expression of these characters. High GCV and PCV were recorded for grain yield per plant and number of effective tillers per plant indicating the variability among the accessions for the traits. Similar results were recorded by Nirmalakumari et al. (2010) and Jyothsna et al. (2016). Heritability estimates were high for grain yield, plant height and number of effective tillers per plant which indicated that these characters were less influenced by environmental conditions and phenotypic selection would be effective. Anuradha et al. (2017) also reported high heritability for these traits. The estimates of broad sense heritability have limitation because it includes both additive and epistatic gene effects. Hence heritability will be more meaningful if combined with genetic advance. High heritability coupled with high genetic advance was observed for grain yield per plant and number of effective tillers per plant indicating the importance of additive gene action in governing the inheritance of these traits. Hence direct selection for these traits is effective for improvement of these traits. Similar findings were reported earlier by Padmaja (1998) for number of effective tillers per plant and grain yield.
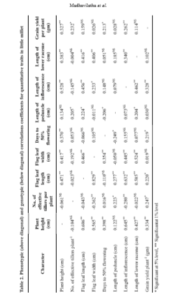
Correlation coefficient measures the mutual relationship between various plant characters and determines the component characters on which selection can be relied upon for genetic improvement of yield. Results of association studies in the study revealed that plant height, length of inflorescence, number of effective tillers per plant, days to 50% flowering and flag leaf length showed high positive and significant correlation with grain yield per plant at both phenotypic and genotypic levels (Table 2). These results are in accordance with findings of Venkataratnam et al. (2019) for number of effective tillers, plant height and length of inflorescence, Shinde et al. (2018), for plant height, length of inflorescence, Suryanarayana and Sekhar (2018) for number of effective tillers, Jyothsna et al. (2016) for days to 50% flowering, Manimozhi et al. (2014) for panicle length and flag leaf length. This suggests selecting for the characters having high positive correlation would improve the grain yield in little millet. In addition, the significant associations between these component traits suggest the possibility of simultaneous improvement of these traits by single selection. Thus selection for these component characters either singly or in combination would be more efficient in improving yield (Nirmalakumari et al. 2010).
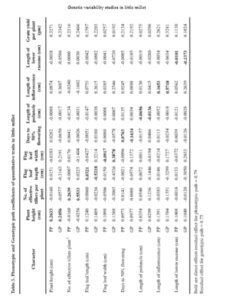
Path analysis of grain yield revealed that plant height, number of effective tillers per plant, length of inflorescence, days to 50% flowering and flag leaf length contributed maximum to the grain yield by having significant positive association and direct effect on grain yield per plant both at genotypic and phenotypic levels (Table 3). Similar findings were reported in little millet were reported by Nandini et al. 2016 for number of effective tillers per plant, panicle length and plant height, Nirmalakumari et al.(2010) for days to 50 per cent flowering, plant height, total number of effective tillers per plant and flag leaf length, Jyothsna et al. (2016) for number of effective tillers per plant days to 50% flowering and flag leaf length showed high positive association with yield even though they have recorded negative direct effect on grain yield at genotypic level. This was due to positive indirect effects via plant height and length of inflorescence.
Madhavilatha et al.
Considering the genetic parameters, character association and direct and indirect effect of various traits, the grain yield improvement is possible through simultaneous improvement of plant height, length of inflorescence, number of effective tillers per plant, days to 50% flowering and flag leaf length.
LITERATURE CITED
Allard, R.W. 1960. Principles of Plant Breeding. John Wiley and Sons Inc., New York, 485.
Anuradha, N., Patro, T.S.S.K., Divya, M., Rani, Y.S and Triveni, U. 2017.Genetic variability, heritability and correlation ofquantitative traits in little millet genotypes. Journal of Pharmacognosy and Phytochemistry. 6(6): 489-492.
Burton, G.W. 1952. Quantitative inheritance in grasses. In: Proceedings of the “6th International Grassland Congress”. 277-283.
Dewey, D.R and Lu, K.H. 1959. A correlation and path co-efficient analysis of components of crested wheatgrass seed production. Agronomy Journal. 51: 515-518.
Falconer, D.S. 1981. Introduction to Quantitative Genetics. 2nd ed. Longman, London. Johnson, H.W., Robinson, H.F and Comstock, H.K. 1955. Estimates of genetic and environmental variability in Soybean. Agronomy Journal. 47: 314-318.
Jyothsna S., Patro T.S.S.K., Rani Y.S., Neeraja, B., Ashok, S. and Triveni, U. 2016. Studies on genetic parameters, character association and path analysis of yield and its components in little millet (Panicum sumatrense).International Journal of Agriculture Sciences 8(5): 1018-1020.
Lush, J.L. 1940. Intra-sire correlation and regression of offspring on dams as a method of estimating heritability of characters. In: Proc. of “American Society of Animal Production”. 33: 293-301.
Manimozhi, V.S., Nirmalakumari, A and Subramanian, A. 2014. Genetics and inter relationships of yield traits for enhancing productivity of little millet. Electronic Journal of Plant Breeding. 5(1): 82- 86.
Nandini, C., Sujata B and Aruna, Y.R.2016. Genetic variability, heritability, genetic advance and character association studies in F3 generation of cross JK8 × Peddasame (purple late) in little millet (Panicum miliare L.). Asian Journal of Bio Science. 11(2): 244-249.
Nirmalakumari, A., Salini, K and Veerabadhiran, P. 2010. Morphological characterization and evaluation of little millet (Panicum sumatrense Roth. ex. Roem. and Schultz.) Germplasm. Electronic Journal of Plant Breeding. 1(2): 148-155.
Padmaja, 1998. Studies on variability, correlation, path analysis and D2 analysis for yield and yield attributes in little millet (Panicum sumatrense Roth.). M.Sc. (Ag.) Thesis. Tamil Nadu Agricultural University, Coimbatore, India.
Sasamala, A.C., Sahoo, L.P., Mohapatra, K.C and Patra, C. 2015. Genetic variability and character association in little millet (Panicum sumatrense) germplasm collections of Odisha, India. Advances in Applied Agricultural Sciences. 3(1): 40-50.
Selvi, M.V., Nirmalakumari, A and Senthil, N. 2015. Genetic diversity for zinc, calcium and iron content of selected little Millet Genotypes. Journal of Nutrition & Food Sciences. 5(6):1-5.
Shinde, S.S., Karad, S.R and Kakde, D.S. 2018. Correlation and path analysis studies in little millet (Panicum sumatrense L.). Green Farming. 9(1): 21-23.
Suryanarayana, L and Sekhar, D. 2018. Studies on genetic variability, character association and path analysis in little millet (Panicum sumatrenseL.) Genotypes. The Pharma Innovation Journal. 7(7): 908-910.
Venkataratnam, T., Madhavilatha, L., ReddiSekhar, M and Nirmal Kumar, A.R. 2019. Studies on correlation and path analysis in little millet (Panicum sumatrense). Green Farming. 10(6): 679-682.
- Genetic Divergence Studies for Yield and Its Component Traits in Groundnut (Arachis Hypogaea L.)
- Correlation and Path Coefficient Analysis Among Early Clones Of Sugarcane (Saccharum Spp.)
- Character Association and Path Coefficient Analysis in Tomato (Solanum Lycopersicum L.)
- Survey on the Incidence of Sesame Leafhopper and Phyllody in Major Growing Districts of Southern Zone of Andhra Pradesh, India
- Effect of Organic Manures, Chemical and Biofertilizers on Potassium Use Efficiency in Groundnut
- A Study on Growth Pattern of Red Chilli in India and Andhra Pradesh

