Studies on Genetic Variability for Yield and Yield Related Traits in Peanut (Arachis Hypogaea L.)
0 Views
ABSTRACT
Thirty three genotypes of peanut were evaluated during kharif 2019 to estimate genetic parameters viz., genetic variance, heritability (broad sense) and genetic advance as per cent of mean for 19 characters. The analysis of variance revealed that highly significant differences were observed among the genotypes for all the characters studied except dry haulms yield per plant. The phenotypic coefficient of variation (PCV) was greater than genotypic coefficient of variation (GCV) for all the characters studied implying that these characters were highly influenced by the environmental effects. High PCV and GCV values (>20%) was recorded for number of primary branches per plant and number of secondary branches per plant. High heritability (> 60%) coupled with high genetic advance as per cent of mean was recorded for specific leaf area at 45 DAS, number of primary branches per plant, hundred pod weight and hundred kernel weight indicating that these characters were governed by additive gene effects and selection of these characters is rewarding
KEY WORDS:
Genetic advance, heritability, variability, peanut.
INTRODUCTION
The cultivated groundnut or peanut (Arachis hypogaea L.), is an annual self pollinated crop with chromosome number of 2n = 4x = 40. It is the only nut found under the soil and designated as “wonder legume”. It belongs to the family Fabaceae. It is native to South America, grown throughout the tropical and sub-tropical regions of the world between the latitudes of 40o N to 40o
S. It is an important oilseed crop known for its several purposes including edible oil production, direct human consumption as food and for animal consumption in the form of silage, hay and oilcake. It is a rich source of high quality oil (44-56%), protein (22-30%) on dry seed basis, carbohydrates (10-25%), vitamins (E, Z and B complex), minerals (Ca, P, Mg, Zn and Fe) and fiber. Being a legume it adds nitrogen and organic matter to the soil.
In India, peanut is cultivated in an area of 48.87 lakh ha with production of 92.52 lakh tonnes and productivity of 1893 kg ha-1. In Andhra Pradesh it is grown in an area of 7.35 lakh ha with production of 10.48 lakh tonnes and productivity of 1426 kg ha-1 (Anonymous, 2018). Hence, increasing the productivity of peanut is one of the prime objective in plant breeding.
Effectiveness of selection depends on presence of considerable genetic variability in gene pool for evolving
desirable plant types. Heritability measures the degree of resemblance between the parents and the off-springs, while genetic advance aids in exercising the necessary selection pressure. Reliable estimates of genetic variability, heritability and genetic advance will be essential for the plant breeders in determining the direction and magnitude of selection.
MATERIAL AND METHODS
Thirty three advanced breeding lines of peanut were sown during kharif, 2019 in a Randomized Block Design (RBD) with three replications in order to study the genetic parameters viz., variability, heritability and genetic advance as per cent of mean. In each replication every genotype was sown in five rows of 5 m length with a spacing of 30 cm between the rows and 10 cm between the plants within the row. The data was collected from five randomly selected plants of each genotype in each replication for 19 characters viz., days to 50% flowering, days to maturity, SPAD chlorophyll meter reading at 45 DAS, specific leaf area at 45 DAS (cm2 g-1), relative water content (%), plant height (cm), number of primary branches per plant, number of secondary branches per plant, number of mature pods per plant, hundred pod weight (g), shelling per cent, hundred kernel weight (g), sound mature kernel per cent, dry haulms yield per plant (g), harvest index (%), protein content (%), oil content (%), kernel yield per plant (g) and pod yield per plant (g).
The various genetic parameters viz., phenotypic coefficient of variance (PCV) and genotypic coefficient of variance (GCV), heritability (h2 ) in broad sense and genetic advance as per cent of mean was calculated as suggested by Burton (1952), Lush (1940), Johnson et al., (1955a) and Johnson et al., (1955b). The data analysis was carried out with WINDOWSTAT 9.2 software.
RESULTS AND DISCUSSION
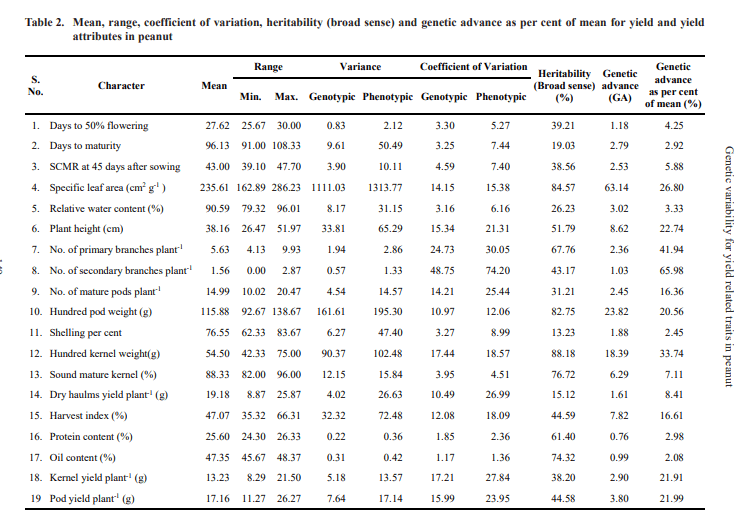
Analysis of variance revealed significant differences were observed for all the characters studied except dry haulms yield per plant indicating the presence of ample amount of variability among the genotypes.(Table 1). The estimates of PCV was higher than GCV for all the characters under study (Table 2 and Fig. 1) indicating
that the characters were influenced by the environment. The characters, number of primary branches per plant (GCV: 24.73%; PCV: 30.05%) and number of secondary branches per plant (GCV: 48.75 %; PCV: 74.20%) registered higher GCV and PCV (>20) values respectively indicating that these characters contributed markedly to the total variability. Similar results were reported by Mahesh et al. (2018), Kamdi et al. (2017), Bhargavi et al. (2017) and Gupta et al. (2015) for number of primary branches per plant and Korat et al. (2009) for number of secondary branches per plant.
Moderate GCV and high PCV was exhibited by kernel yield per plant (GCV: 17.21%; PCV: 27.84%), pod yield per plant (GCV: 15.99%; PCV: 23.95%), plant height (GCV: 15.34%; PCV: 21.31%), number of mature pods per plant (GCV: 14.21%; PCV: 25.44%) and dry haulms yield per plant (GCV: 10.49%; PCV: 26.99%).
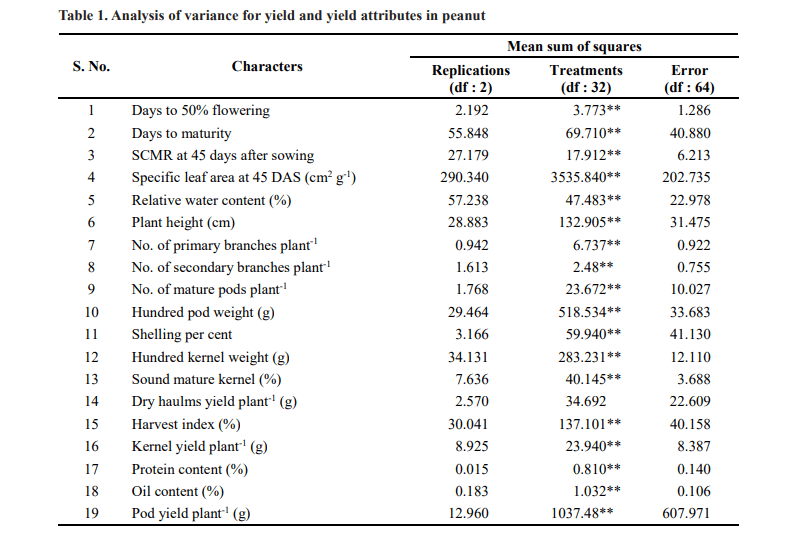
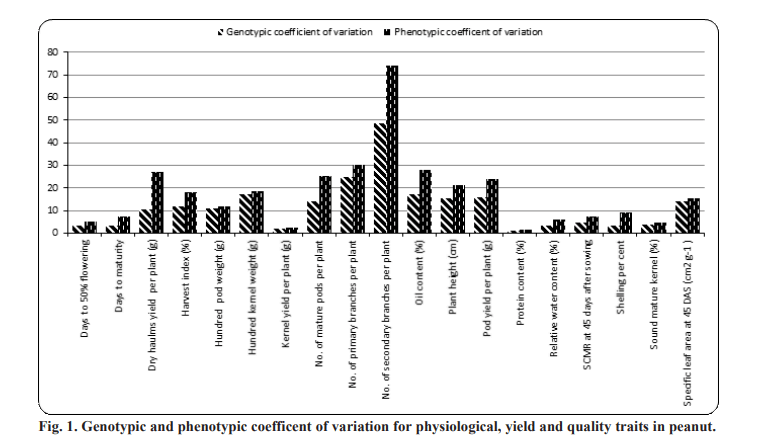
Chavadhari et al. (2017) and Yusuf et al. (2017) reported moderate values of GCV and high PCV for plant height. Moderate GCV and moderate PCV was exhibited by specific leaf area at 45 DAS (GCV: 14.15%; PCV: 15.38
%), harvest index (GCV: 12.08%; PCV: 18.09%), hundred pod weight (GCV: 10.97%; PCV: 12.06%) and hundred kernel weight (GCV: 17.44%; PCV: 18.57%). Moderate variability for hundred kernel weight was reported by Nayak et al. (2018), Nagaveni and Hasan (2019), Rathod and Toprope (2018) and Bhakal and Lal (2017). Similar results for hundred pod weight was reported by Korat et al. (2010) and Mahesh et al. (2018). Moderate variability for harvest index and specific leaf area at 45 DAS was reported by Nagaveni and Hasan (2019).
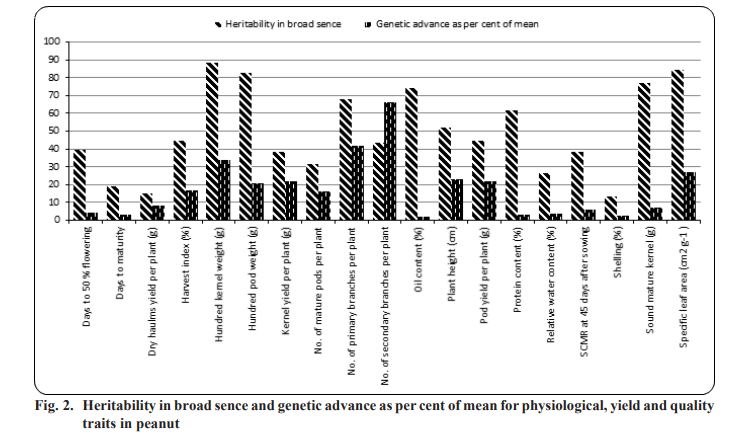
High heritability was recorded by hundred kernel weight (88.18%), followed by specific leaf area (84.57%), hundred pod weight (82.75%), sound mature kernel per cent (76.72%), oil content (74.32%), number of primary branches per plant (67.76%) and protein content (61.40%) indicating that the effect of environment is least in expression of these characters.
Heritability in broad sense includes both additive and epistatic gene effects, it will be reliable only if accompanied by high genetic advance (Fig. 2). High heritability coupled with high genetic advance as per cent of mean were recorded for the characters viz., specific leaf area at 45 DAS (h2bs= 84.57%, GAM= 26.80%), number of primary branches per plant (h2bs= 67.76%, GAM= 41.94%), hundred kernel weight (h2bs= 88.18%, GAM= 33.740%) and hundred pod weight (h2 = 82.75%, GAM= 20.56%) indicating the preponderance of additive gene action in expression of these characters and selection would be effective for improvement of these characters. High heritability coupled with high genetic advance as per cent of mean for number of primary branches per plant was reported by Waidkar et al. (2018), Mahesh et al. (2018) and Vasanthi et al. (2015a). High heritability coupled with high genetic advance as per cent of mean for hundred kernel weight were also reported by Satish (2014), Patil et al. (2014), Rao et al. (2014), Gupta et al. (2015), Vasanthi et al. (2015a), Bhargavi et al. (2017), Mahesh et al. (2018) and Kakeeto et al. (2019). In the present study the estimates for hundred pod weight was in accordance to results reported by Nath and Alam (2002), Gupta et al. (2015), Chavadari et al. (2017) and Kumar et al. (2019).
CONCLUSION
High heritability coupled with high genetic advance as per cent of mean was observed for specific leaf area at 45 DAS, number of primary branches per plant, hundred kernel weight and hundred pod weight indicating the preponderance of additive gene action in governing these traits and direct selection would be rewarding for crop improvement. It was also observed that the trait number of primary branches per plant possessed higher estimates of GCV, PCV, heritability and genetic advance as per cent of mean implying that these traits were predominantly under the control of additive gene action and genetic improvement can be achieved through simple selection for these traits.
ACKNOWLEDGEMENT
The authors thank Acharya N.G. Ranga Agricultural University for providing financial assistance and support in the conduct of experiment at RARS, Tirupati, A.P.
LITERATURE CITED
Anonymous, Annual Report, 2018. All India Coordinated Research Project on Groundnut, Directorate of Groundnut Research. Junagadh, Gujarat.
Bhargavi, G., Rao, V.S and Babu, D.R. 2017. Studies on variability, heritability and genetic advance as per cent of mean in spanish bunch groundnut genotypes (Arachis hypogaea L.). Legume Research. 40(4): 773-777.
Bhakal, M and Lal, G.M. 2017. Estimation of genetic variability, correlation and path analysis in groundnut (Arachis hypogaea L.) germplasm. Chemical Science Reviews and Letters. 6(22): 1107-1112.
Burton, G.W. 1952. Quantitative inheritance in grasses. Proceedings of Sixth International Grassland Congress. 1: 277-283.
Chavadhari, R.M., Kachhadia, V.H., Vacchani, J.H and Virani, M.B. 2017. Genetic variability studies in groundnut (Arachis hypogaea L.) Electronic Journal of Plant Breeding. 8(4): 1288-1292.
Nayak, P.G., Venkataiah, M., Revathi, P and Srinivas, B. 2018. Correlation and genetic variability studies in groundnut (Arachis hypogaea L.) genotypes. International Journal of Genetics. 10(2): 354-356.
Gupta, R.P., Vachhani, J.H., Kachhadia, V.H., Vaddoria,
M.A and Reddy, P. 2015. Genetic variability and heritability studies in Virginia groundnut (Arachis hypogaea L.). Electronic Journal of Plant Breeding. 6(1): 253-256.
Johnson, H.W., Robinson, H.F and Comstock, R.E. 1955a. Estimates of genetic and environmental variability in soybean. Agronomy Journal. 47: 413-418.
Johnson, H.W., Robison, H.F and Comstock, R.E. 1955b. Genotypic and phenotypic correlations in soybean and their implications in selection. Agronomy Journal. 47: 477-83.
Kamdi, S.R., Shanti, P.R., Kadu, P.R., Shubangi, B.P and Kothikar, R.B. 2017. Genetic variability, correlation and path analysis in groundnut. BIOINFOLET – A Quarterly Journal of Life Sciences. 14(1): 36-39.
Kakeeto, R., Baguma, S.D., Subire, R., Kaheru, J., Karungi, E and Biruma, M. 2019. Genetic variation and heritability of kernel physical quality traits and their association with selected agronomic traits in groundnut (Arachis hypogeae L.) genotypes from Uganda. African Journal of Agricultural Research. 14(10): 597-603.
Korat, V.P., Pithia, M.S., Savaliya, J.J., Pansuriya, A.G and Sodavadiya, P.R. 2009. Studies on genetic variability in different genotypes of groundnut (Arachis hypogaea L.). Legume Research. 32(3): 224-226.
Korat, V.P., Pithia, M.S., Savaliya, J.J., Pansuriya, A.G and Sodavadiya, P.R. 2010. Studies on characters association and path analysis for seed yield and its components in groundnut (Arachis hypogaea L.). Legume Research. 33(3): 211-216.
Kumar, N., Ajay, B.C., Rathanakumar, A.L., Radhakrishnan, T., Mahatma, M.K., Kona, P and Chikani, B.M. 2019. Assessment of genetic variability for yield and quality traits in groundnut genotypes. Electronic Journal of Plant Breeding. 10(1): 196-206.
Lush, J.L. 1940. Inter size correlation and regression of offspring on dams as a method of estimating heritability of characters. Proceedings of American Society of Animal Production. 33: 293-301.
Mahesh, R.H., Hasan, K., Temburne, B.V., Janila, P and Amaregouda, A. 2018. Genetic variability,
correlation and path analysis studies for yield and yield attributes in groundnut (Arachis hypogaea L.). Journal of Pharmacognosy and Phytochemistry. 7(1): 870-874.
Nath, U.K and Alam, M.S. 2002. Genetic variability, heritability and genetic advance of yield and related traits of groundnut. Journal of Biological Science. 2(1): 762-764.
Nagaveni and Hasan, K. 2019. Genetic variability studies in terminal drought tolerant groundnut (Arachis hypogaea L.). Journal of Pharmacognosy and Phytochemistry. 9(2): 747-750.
Patil, A.S., Punewar, A.A., Nandanwar, H.R and Shah,
K.P. 2014. Estimation of variability parameters for yield and its component traits in groundnut (Arachis hypogaea L.). The Bioscan. 9(2): 749-754.
Rao, V.T., Venkanna, V., Bhadru, D and Bharathi, D. 2014. Studies on variability, character association and path analysis on groundnut (Arachis hypogaea L.). International Journal of Pure and Applied Bioscience. 2(2): 194-197.
Rathod, S.S and Toprope, V.N. 2018. Studies on variability, character association and path analysis in Groundnut (Arachis hypogaea L.). International Journal of Pure Applied Biosciences. 6(2): 1381- 1388.
Satish, Y. 2014. Genetic variability and character association studies in groundnut (Arachis hypogaea L.). International Journal of Plant, Animal and Environmental Sciences. 4(4): 298-300.
Vasanthi, R.P., Suneetha, N and Sudhakar, P. 2015a. Genetic variability and correlation studies for morphological, yield and yield attributes in groundnut (Arachis hypogaea L.). Legume Research. 38(1): 9-15.
Wadikar, P.B., Dake, D.A., Chavan, M.V and Thorat, G.S. 2018. Character association and variability studies of yield and Its attributing characters in groundnut (Arachis hypogaea L.). International Journal of Agricultural Science and Research. 6: 924-929.
Yusuf, Z., Zeleke, H., Mohammed, W., Hussein, S and Hugo, A. 2017. Correlation and path analysis of groundnut (Arachis hypogaea L.) genotypes in Ethiopia. International Journal of Plant Breeding and Crop Science. 4(2): 197-204.
- Effect of Sowing Window on Nodulation, Yield and Post – Harvest Soil Nutrient Status Under Varied Crop Geometries in Short Duration Pigeonpea (Cajanus Cajan L.)
- Nanotechnology and Its Role in Seed Technology
- Challenges Faced by Agri Startups in Andhra Pradesh
- Constraints of Chcs as Perceived by Farmers in Kurnool District of Andhra Pradesh
- Growth, Yield Attributes and Yield of Fingermillet (Eleusine Coracana L. Gaertn.) as Influenced by Different Levels of Fertilizers and Liquid Biofertilizers
- Consumers’ Buying Behaviour Towards Organic Foods in Retail Outlets of Ananthapuramu City, Andhra Pradesh

