Morphological Characterization of Sesame (Sesamum Indicum L.) Genotypes
0 Views
NAGABHUSHANA*, D. BHARATHI, P. SRIVALLI, B. RUPESH KUMAR REDDY AND M. REDDI SEKHAR
Department of Genetics and Plant Breeding, S.V. Agricultural College, ANGRAU, Tirupati-517 502.
ABSTRACT
One hundred sesame (Sesamum indicum L.) genotypes were evaluated for twenty morphological traits using DUS descriptors in an Alpha Lattice Design with two replications during the rabi season 2024–2025 at the Regional Agricultural Research Station, Tirupati. Based on the results of frequency distribution, a majority of sesame accessions were found to possess medium duration of 50% flowering with light purple petal colour, dense petal hairiness, medium plant branching, medium plant height, basal branching pattern, absent stem hairiness, medium leaf size, slightly lobed leaf, strong leaf serration, absent capsule hairiness, four locules per leaf axil, broad oblong capsule shape, one capsule per leaf axil, alternate capsule arrangement, medium maturity, medium thousand seed weight, white seed coat colour and low oil content percent. The study highlights distinct morphological variations among sesame genotypes due to genetic differences, offering valuable insights for breeders in varietal selection, identification, and conservation.
KEYWORDS: DUS testing, Sesame, Genotypes.
INTRODUCTION
Sesame (Sesamum indicum L.) belongs to the family Pedaliaceae, having ploidy level 2n = 26 is considered as valuable oilseed crop at global level. The seeds contain substantial amount of oil (45–55%), protein (18–25%), carbohydrates (16–18%), fibers and phenolic compounds. It also contains significant amounts of mineral nutrients (Bharathi et al., 2015) along with antioxidants like sesamin and sesamolin.
Globally, sesame is cultivated in an area of 128.36 lakh hectares with a production of 67.41 lakh tonnes and productivity of 525 kg/ha. In India, it covers 10.39 lakh hectares, producing 4.29 lakh tonnes at 413 kg/ha (Indiastat, 2023–2024), highlighting the need for high- yielding varieties suited for diverse agroclimatic regions.
However, challenges such as overlapping local names, inconsistent documentation, and phenotypic similarities among accessions frequently lead to identity issues, including duplication and mislabelling within germplasm collections (Upadhyay et al., 2010). To address these problems, morphological characterization using DUS (Distinctness, Uniformity, and Stability) descriptors offers a standardized and reliable method (PPV&FRA, 2007). Using specific qualitative and quantitative traits helps in clearly identifying and classifying genotypes. Therefore, the present study was undertaken to characterize the released varieties of sesame using DUS descriptors.
MATERIAL AND METHODS
100 genotypes were evaluated in Alpha lattice Design (ALD) with two replications during rabi season 2024-2025 at the Regional Agricultural Research Station in Tirupati. Each genotype was sown in 2 rows of 3-meter length with spacing of 30 cm × 15 cm. The crop was raised under recommended package of practices along with prophylactic protection measures. The observations were recorded on days to 50% flowering, petal colour, petal hairiness, plant height, plant branching, branching pattern, stem hairiness, leaf lobes, leaf size, leaf serration, capsule hairiness, locule number per capsule, capsule shape, capsule number per leaf axil, capsule arrangement, capsule length, days to maturity, seed coat colour, thousand seed weight and oil content.
DUS Testing: Twenty morphological descriptors (PPV&FRA, 2007) have been considered essential for the description of 100 genotypes of sesame using guidelines for the conducting test for distinctiveness, uniformity and stability in sesame.
RESULTS AND DISCUSSION
Based on variation in physical characteristics, it was attempted to group the sesame genotypes and identify each and every one of them through descriptors. Based on morphological variation, the 100 genotypes could be identified from each other. The results for each trait are described briefly presented in Table 1.
Table 1 Morphological characterization of genotypes for descriptor exhibition with frequency
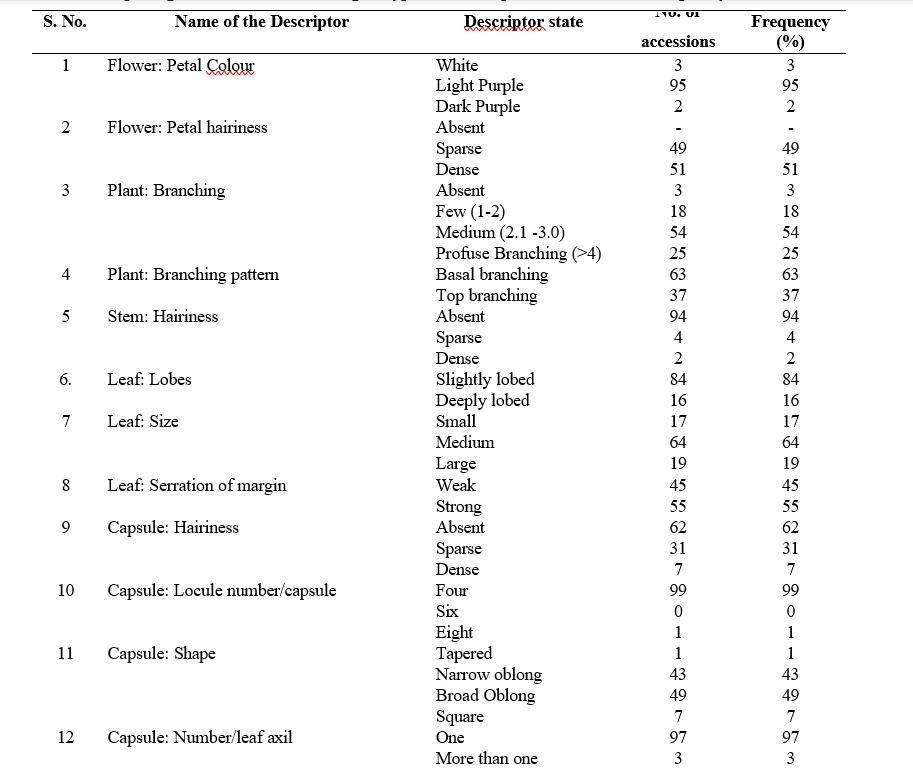


Based on days to 50 per cent flowering, the genotypes were grouped as 64 medium (36-45 days), two early (<36 days) and 34 genotypes late (>45 days) duration of flowering. Three groups were made based on petal colour of flower. Out of the 100 genotypes, 95 had light purple, 3 had white, and 2 had dark purple petal colour. Based on petal hairiness, genotypes were grouped as sparse (49 genotypes) and dense (51 genotypes).
The height of the main stem, a key trait for distinguishing sesame genotypes, showed considerable variability. Based on stem height, genotypes were categorized six genotypes as short (<75 cm), 89 genotypes medium (75–125 cm) and five genotypes as tall (>125 cm). Variation was also observed in the number of primary branches per plant: three genotypes had no branches, 18 had few (1–2), 54 had medium (2.1–4), and 25 had profuse branching (>4). Based on branching pattern the genotypes were grouped as 63 with basal and remaining 37 with top branching pattern.
Among 100 genotypes, four genotypes exhibited sparse stem hairiness, ninety-four genotypes exhibited absence of hair and two genotype exhibited dense hairiness Similar findings and grouping of genotypes based on flower and stem morphological characters were made by Bhoot et al. (2019), Frary et al. (2015) in sesame.
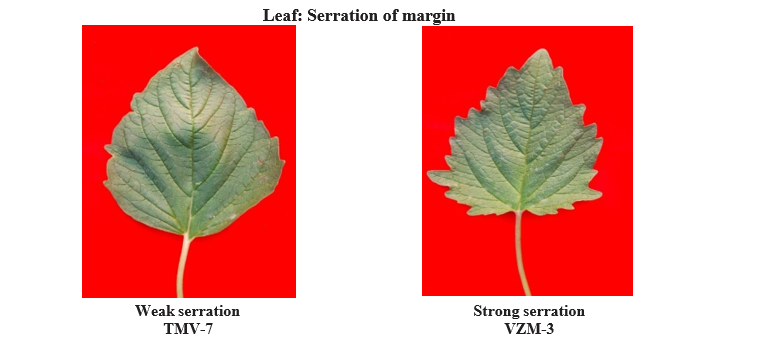
Based on leaf lobes, the genotypes differentiated into two groups. 84 genotypes were having slightly lobed leaves and 16 genotypes were deeply lobed. For leaf size genotypes were classified into three categories, 17 genotypes had small leaf, 64 with medium leaves and remaining 19 genotypes showed large leaf. Difference was also found in leaf serration of margin, 55 genotypes showed strong and remaining 45 genotypes showed weak margin. For capsule hairiness among the genotypes, 62 recorded as absent hairiness, 31 recorded as sparse and the remaining seven genotypes recorded as dense hairiness types. Similarly, the variation was found among the genotypes for number of locules per capsule, 99 genotypes had four locules per capsule, only one had eight locules per capsule.
Four groups were made based on shape of capsule. Out of 100 genotypes, one genotype showed tapered,
43 genotypes showed narrow oblong, 49 genotypes
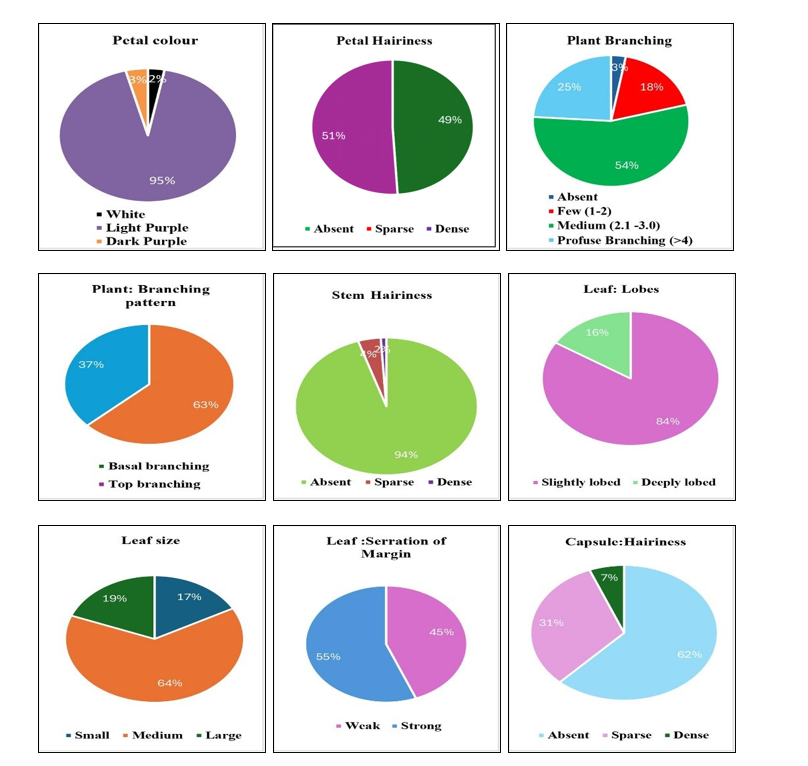
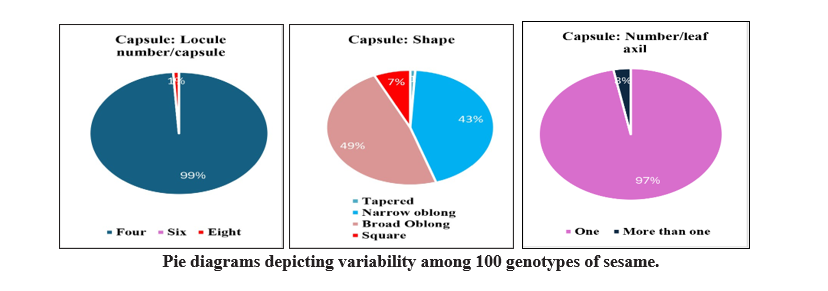
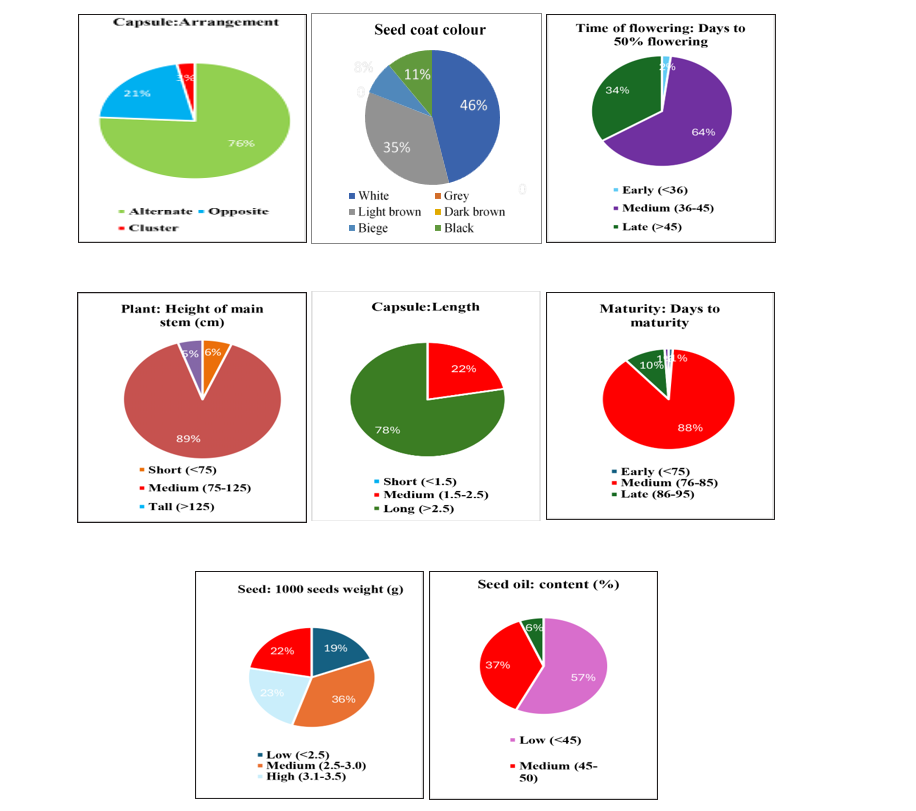
recorded as broad oblong types and seven were square type. The study of number of capsules per leaf axil were grouped into two classes. Viz., 97 genotypes had one and remaining three genotypes showed more than one capsules per leaf axil. Difference was also observed in arrangement of capsule, 76 genotypes were found alternate, 21 genotypes found opposite and remaining three genotypes found cluster types. Two groups were made based on length of capsule i.e long and medium. Out of 100 studied genotypes for length of the capsule, 22 genotypes were showed medium and 78 genotypes observed long length of capsule. Similar observations and grouping of genotypes based on capsule morphological characters were made by Palakshappa et al. (2020), Narayanan and Murugan (2013) in sesame.
Days to maturity in sesame helps to find the genotypes with environmental suitability and avoiding stress conditions. Out of 100 genotypes one genotype showed early, 88 genotypes showed medium, ten


genotypes were late in maturity and only one genotype exhibited very late days in maturity. A wide range of variation is observed for seed coat colour, 54 genotypes exhibited white colour, 8 genotypes had beige colour, 30 genotypes were light brown colour and 8 genotypes recorded black seed colour. In case of 1000-seed weight, 19 genotypes recorded low, 36 genotypes with medium, 23 genotypes recorded high and remaining 22 genotypes showed very high 1000 seed weight. Oil content (OC) percent is a complex quantitative trait and is affected by genotype and environment. 57 genotypes with low (>45 %), 37 genotypes (37 %) with medium (45-50 %) and remaining 6 genotypes (6 %) with high oil content (>50 %). Similar findings were observed by Pavani et al. (2020), Vanishree et al. (2022).
Based on the above results it is clear that profound variation is present among genotypes. This variation is useful for genotype identification and helps in future breeding purpose to produce high yielding genotypes using trait specific germplasm.
LITERATURE CITED
Bharathi, D., Rao, V.T., Venkanna, V. and Bhadru, D. 2015. Association analysis in sesame (Sesamum indicum L.) International Journal of Applied Biology and Pharmaceutical Technology. 6 (1): 210-212
Bhoot, H.V., Sharma, L.K., Kulkarni, G.U., Ravat, U and Rathva, S. 2019. Characterization of sesame genotypes through morphological characters. Journal of Pharmacognosy and Phytochemistry. 8(3): 3132-3138.
Frary A, Tekin P, Celik I, Furat S, Uzun B, Doganlar 2015. Morphological and molecular diversity in sesame germplasm and core set selection. Crop Science. 55:702-711.
Narayanan R and Murugan S.2013 Studies on variability and heritability in sesame (Sesamum indicum L.). International Journal of Current Agricultural Research 2013; 2(11):52-55.
Palakshappa, M.G., Banu, H., Parmeshwarappa, S.G., Bisen, R., Nagappa, H and Holeyannavar, P. 2020. DUS testing of sesame (Sesamum indicum L.) Accessions using morphological descriptors and evaluation for foliar diseases of sesame. International Journal of Current Microbiology and Applied Sciences. 9(1): 1837-1852.
Pavani, K., Lal Ahamed, M., Ramana, J.V and Sirisha, A.B.M., 2020. Studies on genetic variability parameters in sesame (Sesamum indicum L.). International Journal of Chemical Studies. 8(4): 101-104.
PPV&FRA (2007). Guidelines for the conduct of test for distinctiveness, uniformity and stability on sesame (Sesamum indicum L.). Protection of Plant Varieties and Farmers’ Rights Authority, Ministry of Agriculture, Government of India.
Upadhyaya, H. D., Yadav, D., Dronavalli, N., Gowda, L. L. and Singh, S. 2010. Mini core germplasm collections for infusing genetic diversity in plant breeding programs. Electronical Journal Plant Breeding 1 (4): 1294-1309.
Vanishree, S.G., Banu, H., Palakshappa, M.G and Holeyannavar, P. 2022. Morphological characterization of sesame (Sesamum indicum L.) germplasm accessions with DUS. The Pharma Innovation Journal.11(12):938-945.
- Effect of Sowing Window on Nodulation, Yield and Post – Harvest Soil Nutrient Status Under Varied Crop Geometries in Short Duration Pigeonpea (Cajanus Cajan L.)
- Nanotechnology and Its Role in Seed Technology
- Challenges Faced by Agri Startups in Andhra Pradesh
- Constraints of Chcs as Perceived by Farmers in Kurnool District of Andhra Pradesh
- Growth, Yield Attributes and Yield of Fingermillet (Eleusine Coracana L. Gaertn.) as Influenced by Different Levels of Fertilizers and Liquid Biofertilizers
- Consumers’ Buying Behaviour Towards Organic Foods in Retail Outlets of Ananthapuramu City, Andhra Pradesh

