Molecular Characterization of Phyhalltoplasmas Associated With Little Leaf Disease of Brinjal (Solanum Melongena L.) In Chittoor District of Andhra Pradesh
0 Views
GOPISETTY SANDHYA*, NEELATURU CHANDRASHEKAR MAMATHA, VASANTHU SRILATHA, PANYAM KARTHIK REDDY AND MADEM GURIVI REDDY
Department of Plant Pathology, S.V. Agricultural College, ANGRAU, Tirupati-517502.
ABSTRACT
Brinjal little leaf (BLL) is a prevalent phytoplasma disease in India that induces severe economic losses every year. During
2021, symptoms of little leaf and witches’ broom were observed on brinjal plants grown in the horticultural fields of S. V. Agri- cultural College, Tirupati. Four symptomatic and two symptomless plants collected from the surveyed places were subjected to genomic DNA extraction and used as template in nested PCR assays with universal phytoplasma 16S rRNA primers, P1/P7 and R16F2n/R16R2. Amplification of ~1.25 kb product was obtained only from symptomatic BLL plants and the positive control, but not from any asymptomatic plants.
KEYWORDS: Eggplant/Brinjal, 16S rRNA, Nested PCR assay.
INTRODUCTION
Brinjal (Solanum melongena L.) is the most important vegetable cultivated all over the world. India is the second largest producer of brinjal in the world and Andhra Pradesh is a leading brinjal growing state of India (Anonymous, 2019). The crop is challenged by several diseases caused by fungi, bacteria, viruses and other insect pests of which brinjal little leaf (BLL) is one of the most important disease caused by phytoplasma causing considerable economic losses as the infected plant fails to produce a single fruit. Phytoplasma associated diseases in vegetable crops are prevalent in Asian, African, and American continents. So far twenty- eight vegetable species have been reported to be infected by different strains of phytoplasma all around the world (Kumari et al., 2019). Brinjal Little Leaf disease was first recorded by Thomas and Krishnaswami (1939) in India with 100% yield loss (Rao and Kumar, 2017). Five ribosomal groups (16SrI, 16SrII 16SrVI, 16SrIX and 16SrXII) were reported associated with BLL disease at global level (Kumari et al., 2019). The phytoplasma groups 16SrI and 16SrVI (subgroup 16SrVI-D) are the most dominant reported to be associated with BLL disease in eggplants showing little leaf, shortening of internodes, witches’ broom, phyllody, stunting and yellowing with necrosis symptoms in India (Kumar et al., 2017; Rao and Kumar, 2017). The phytoplasma disease of eggplant was reported to be widespread in the areas, where overlapping crop cycles and weeds ensure high populations of leafhoppers and provide natural reservoirs for the different strains of phytoplasma (Rao and Kumar, 2017; Rao, 2021). But limited reports are available from Andhra state of India. Since, most of the farmers in Andhra Pradesh are cultivating brinjal for commercial sale in local markets, an attempt was made to detect, phytoplasma presence associated with BLL in the Chittoor district of Andhra Pradesh.
MATERIAL AND METHODS
During April 2021 survey was conducted in horticultural fields of S. V. Agricultural College, Tirupati, Chittoor district, Andhra Pradesh. The disease incidence of BLL was assessed by counting number of infected plants visually over healthy plants by randomly selecting 5 × 5 meter plot at each location. Moreover symptomatic leaf samples were collected in each surveyed location were brought to the laboratory along with two healthy leaf samples for further processing. DNA was extracted from the symptomatic and non-symptomatic brinjal samples using CTAB method.
DNA extraction
The total genomic DNA was isolated from the leaves of healthy and symptomatic plants by following CTAB (Cetyl Trimethyl Ammonium Bromide) method of Murray and Thomson (1980) with some modifications. The modifications were made to improve the quality of DNA.
Procedure
- The leaf samples were grounded into a fine powder with liquid nitrogen.
- Approximately 100 mg of the powder was transferred into 2 ml eppendorf tube by spatula, to this 5 ml of CTAB extraction buffer was added and then incubated in water bath at 65°C for 45 min to 1 hr with occasional vortexing at every 20 min interval.
- The tubes were removed from the water bath and allowed to cool at room temperature and later centrifuged at 14,000 rpm for 10 min.
- Then equal volume of Phenol: Chloroform (1:1 v/v) was added to the supernatant and mixed thoroughly by gentle inversion and centrifuged for 20 min at 13,000 rpm at 25°C and then the supernatant was carefully pipetted out into new 2 ml eppendorf
- To this supernatant, chloroform: Isoamyl alcohol (24:1 v/v) was added then mixed thoroughly by gentle inversion and centrifuged at 12000 rpm for 15 min at 25°C. Then supernatant was carefully pipetted out into new 1.5 ml eppendorf tubes.
- The supernatantwas taken to which ice cold isopropanol of about 0.6 volumes (2/3rd of pipetted volume) and 1volume of 3M sodium acetate of pH 5.2 was added. The contents were mixed gently by inversion and kept undisturbed for overnight or for 2 hrs at -20°C.
- Subsequently, the tubes were centrifuged at 13,000 rpm for 20 min at 4°C temperature to pellet out the
- The supernatant was discarded gently and the DNA pellet was washed with 70% ethanol and centrifuged at 13,000 rpm for 10 minutes.
- The supernatant was removed and the tubes were allowed to air dry completely until ethanol smell was lost and then the pellet was dissolved in 40-80 μl of nuclease free water.
Quality and Quantity of DNA
DNA was assessed for its purity and intactness using agarose gel electrophoresis.
1. Quantification of DNA by 1% Agarose Gel Electrophoresis
Preparation of 1% Agarose Gel: 1 g of agarose was placed in a conical flask containing 100 ml 1X TAE buffer. The conical flask along with its contents was placed in an oven until agarose gets melted completely
and clear solution was formed and then the flask was taken out from the oven and allowed to cool. 3 µl of Ethidium Bromide (10 mg ml-1) was added to this 100 ml of agarose gel and mixed thoroughly. Later the solution was poured slowly into the gel casting tray which is pre- set with 0.5 mm combs, to avoid the formation of bubbles. After solidification the gel with casting tray was placed in gel tank and the comb was removed gently without disturbing the wells that formed upon solidification.
2) Quantification of DNA by NanoDrop spectrophotometer
The NanoDrop spectrophotometer (model ND1000) was used to assess the quantity and quality of DNA employing the following procedure. Before initializing the NanoDrop Reader, the pedestal was cleaned with tissue paper to remove the dust particles. Then for initializing the instrument, 1-2 μl of distilled water was placed on the lower pedestal, closed the upper one and clicked on measure option. Then the pedestal was cleaned with tissue paper and 1.5 μl of 1X TE buffer was placed on lower pedestal and repeated the procedure for blank measurement. After that, 2 μl of DNA sample was placed to measure the quality and quantity at A260 nm and A280 nm to assess the purity of DNA. The process is repeated for all the DNA samples. A ratio of ~1.8 is generally accepted as “pure” for DNA; a ratio of ~2.0 is generally accepted as “pure” for RNA. If the ratio is lower in either case, it may indicate the presence of protein, phenol or other contaminants that absorb strongly at/or near 280 nm.
Normalization of DNA concentration
Normalization of DNA samples was done to equalize the concentration of all the samples to be used for PCR reaction. The purpose of normalization was to avoid erroneous analyses due to difference in the brightness of the bands obtained after electrophoresing the amplified PCR products. Normalization was done by diluting the DNA samples with sterile distilled water to their required dilution factor which in turn depends upon the initial concentration of DNA sample (from quantification readings) and the type of analysis done (markers used). After normalization of samples the concentration of DNA was 100 ng µl-1.
It was done by using the formula: N1V1 = N2V2 PCR Amplification of Phytoplasmas DNA Primers used for 16S rRNA gene
The extracted DNA from the plants and insects was amplified for 16S ribosomal DNA with phytoplasma specific universal primer pair P1/P7 (Deng and Hiruki, 1991; Schneider et al., 1995) followed by nested primer
Table 1. Details of Primers used for amplification of phytoplasma DNA in collected sample
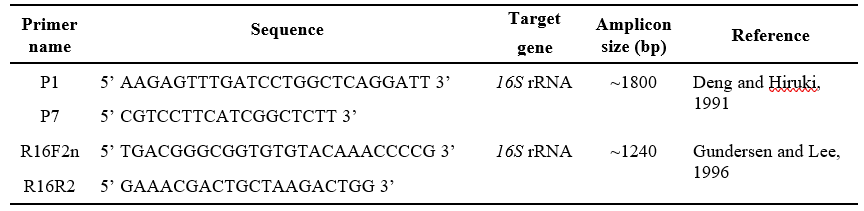
pair R16F2n/R16R2 (Gundersen and Lee, 1996) (Table 1).
PCR cycles used for amplification
PCR reactions were carried out in a Mastercycler (Eppendorf, Germany) and the cycling protocols for the amplification of different genes used are described herein.
16S rRNA gene
First round of PCR assay for the conserved region of the 16S rRNA gene using P1/P7 primer pair and the cycling protocol used in the study are mentioned here under (Table 2).
Table 2. PCR programme for amplification of 16S rDNA (first round PCR)
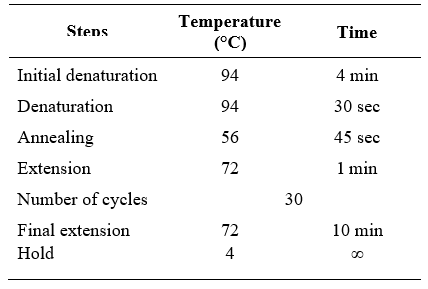
For nested PCR amplification of 16S rRNA using R16F2n/R16R2 primer pair and the cycling protocol used are mentioned in Table 3.
RESULTS AND DISCUSSION
From all the collected samples, the affected brinjal plants showed the symptoms of littleleaf and witches’ broom (Figures 1a and 1b). The disease incidence was recorded from10% to 20%. The association of phytoplasmas in all the symptomatic samples was
Table 3. PCR programme for amplification of 16S rDNA (second round PCR)

confirmed by PCR amplification of ~1.8 kb products in the first round (Figure 2a). PCR and ~1.2 kb products in nested PCR assays with primer pair P1/P7 and R16F2n/R16R2, respectively, while no amplification was observed in the asymptomatic samples (Figure 2b) which confirmed the association of phytoplasma with symptomatic brinjal plants.
DISCUSSION
Phytoplasma belonging to 16SrII-D subgroup was found as the most widely distributed phytoplasma strain on other plant crops in India (Rao, 2021; Reddy et al., 2021) . Earlier the 16 srII- D phytoplasm were identified on brinjal from Uttar Pradesh and Karnataka (Kumar et al., 2017; Yadav et al., 2016) and 16SrVI-D (Azadvar and Baranwal, 2012; Kumar et al., 2017) was reported from various parts of India whereas, the presence of 16SrVI-D and 16SrI group in Bangladesh (Siddique et al., 2001), 16SrII-A in China (Cai et al., 2016), 16SrII-D in Oman (Al-Subhi et al., 2018) was reported in BLL diseased plants.
Naik et al. (2018) reported the association of 16SrVI-D sub-group of phytoplasma in Andhra Pradesh.


Lane M: 1kb ladder, Lane N: Negative control, Lane P: Positive control,
Lane 1: brinjal isolate sample-1 and Lane 2- Brinjal isolate sample-2
Figure 2a. Gel electrophoresis image for direct PCR assay results of phytoplasma showing expected amplicons from symptomatic brinjal plants obtained with primer pairs P1/P7.
In the present study, the association of 16SrII-D from Andhra Pradesh was confirmed based on sequence comparison, phylogeny and RFLP analysis of 16S rDNA sequences. To our knowledge, this is the first report of the showing the association of 16SrII-D sub-group of phytoplasma with little leaf of brinjal in Andhra Pradesh. Recently, 16SrII-D subgroup phytoplasma was identified and characterized from chickpea and weeds from Kadapa, Andhra Pradesh (Reddy et al., 2021). In India, brinjal is cultivated in the same season with many other agricultural crops. The scenario of natural phytoplasma spread from brinjal to other plant species and vice versa, through an efficient vector species, is quite possible as reported in other states of India (Kumar et al., 2017).

Lane M: 1 kb ladder, Lane N: Negative, Lane P: Positive control,
Lane 1: Brinjal isolate sample-1, Lane 2- Brinjal isolate sample-2
Figure 2b. Gel electrophoresis image for nested PCR assay results of phytoplasma showing expected amplicons from symptomatic sesame plants obtained with primer pairs R16F2n/R16R2.
This indicates that Brinjal Little Leaf disease is widely spreading and association of new sub-groups were reported based on the prevalence of alternate hosts and vectors in a particular locality. Keeping this in view, further studies on epidemiology along with possible natural spread sources and management are therefore a foremost need.
LITERATURE CITED
Al-Subhi, A.M., Hogenhout, S.A., Al-Yahyai, R.A and Al-Sadi, A.M. 2018. Detection, identification, and molecular characterization of the 16SrII-D phytoplasmas infecting vegetable and field crops in Oman. Plant Disease. 102: 576-588.
Anonymous 2019. http://www.mospi.gov.in/ [accessed 02.06.2021]
Azadvar, M and Baranwal, V.K. 2012. Multilocus sequence analysis of phytoplasma associated with brinjal little leaf disease and its detection in Hishimonas phycitis in India. Phytopathogenic Mollicutes. 2: 15-21.
Cai. H., Wang, L., Mu, W., Wan, Q., Wei, W., Davis, R.E., Chen, H and Zhao, Y. 2016. Multilocus genotyping of a ‘Candidatus Phytoplasma aurantifolia’-related strain associated with cauliflower phyllody disease in China. Annals of Applied Biology. 169. 64-74.
Deng, S and Hiruki, C. 1991. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. Journal of Microbiological Methods. 14: 53-61.
Gundersen, D.E and Lee, I.M. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathologia Mediterrenea. 35: 144-151.
Kumar, M., Madhupriya and Rao, G.P. 2017. Molecular characterization, vector identification and sources of phytoplasmas associated with brinjal little leaf disease in India. 3 Biotech. 7: 1-11.
Kumari, S., Nagendran, K., Rai, A.B., Singh, B., Rao,
G.P and Bertaccini, A. 2019. Global status of phytoplasma diseases in vegetable crops. Frontiers in microbiology. 10: 1349.
Murray, M.G and Thompson, W.F. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 8: 4321-5.
Naik, D.V.K., Reddy, B.V.B., Rani, J.S., Devi, S.J.R
and Prasad, K.V.H. 2018. Candidatus Phytoplasma trifolii associated with little leaf disease of Solanum melongena (Brinjal) in Andhra Pradesh, India. Journal of Pharmacognosy and Phytochemistry. 7: 3695-3697.
Rao, G.P. 2021. Our understanding about phytoplasma research scenario in India. Indian Phytopathology. 1-31.
Rao, G.P and Kumar, M. 2017. World status of phytoplasma diseases associate with eggplant. Crop Protection. 96: 22-29.
Reddy, M.G., Baranwal VK, Sagar, D and Rao, G.P. 2021. Molecular characterization of chickpea chlorotic dwarf virus and peanut witches’ broom phytoplasma associated with chickpea stunt disease and identification of new host crops and leafhopper vectors in India. 3 Biotech. 11(3): 1-23.
Schneider, B., Seemüller, E., Smart, C.D and Kirkpatrick, B.C. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas. In: Molecular and Diagnostic Procedures in Mycoplasmology. Vol. 2, pp 369–380. Eds S Razin and JG Tully. Academic Press,
Siddique, A.B.M., Agrawal, G.K., Alam, N and Reddy,M.K 2001. Electron microscopy and molecular characterization of phytoplasmas associated with little leaf disease of brinjal (Solanum melongena L.) and periwinkle (Catharanthus roseus) in Bangladesh. Journal of Phytopathology. 149: 237- 244.
Thomas, K.M and Krishnaswami, C.S. 1939. Little leaf—a transmissible disease of brinjal. Proceedings of the National Academy of Sciences. 10: 201-212.
Yadav, V., Mahadevakumar. S., Tejaswini, G.S., Shilpa, N., Amruthavalli and Janardhana, G.R. 2016. First report of 16SrII-D phytoplasma associated with eggplant big bud (Solanum melongena L.) in India. Plant Disease. 100: 517.
- Bio-Formulations for Plant Growth-Promoting Streptomyces SP.
- Brand Preference of Farmers for Maize Seed
- Issues That Consumer Experience Towards Online Food Delivery (Ofd) Services in Tirupati City
- Influence of High Density Planting on Yield Parameters of Super Early and Mid Early Varieties of Redgram (Cajanus Cajan (L.) Millsp.)
- Influence of Iron, Zinc and Supplemental N P K on Yield and Yield Attributes of Dry Direct Sown Rice
- Effect of Soil and Foliar Application of Nutrients on the Performance of Bold Seeded Groundnut (Arachis Hypogaea L.)

