Genetic Variability Studies for Quantitative Traits and Its Components in Inbred Lines of Pearl Millet (Pennisetum Glaucum (L.) R. BR.)
0 Views
I. PRAVEEN KUMAR*, M. SHANTHI PRIYA, L. MADHAVILATHA AND P. LATHA
Department of Plant Pathology, S.V. Agricultural College, ANGRAU, Tirupati-517 502.
ABSTRACT
In the present study 70 pearl millet inbred lines were evaluated for 21 quantitative traits. Analysis of variance for 70 pearl
millet inbred lines revealed significant differences among the germplasm lines for all the 21 traits studied indicating the presence of considerable genetic variability among the lines studied. The high estimates of GCV and PCV recorded for grain yield plant-1 followed by 1000 grain weight, panicle weight, green fodder yield plant-1, dry fodder yield plant-1 and harvest index indicated the presence of high genetic variability for these characters. High heritability coupled with high genetic advance as per cent of mean was recorded for days to 50% flowering, spike length, spike girth, number of productive tillers plant-1, plant height, 1000 grain, panicle weight, green fodder yield plant-1, dry fodder yield plant-1, threshing percentage, harvest index and grain yield plant-1 indicating the preponderance of additive gene action. These characters can be further improved by following simple selection procedure.
KEYWORDS: Pearl millet, PCV, GCV, Heritability, Genetic advance.
INTRODUCTION
Pearl millet (Pennisetum glaucum (L.) R. Br., 2n = 14) is an annual, C4 cereal crop with high photosynthetic efficiency and belongs to the family Gramineae. It has its origin in central tropical Africa and is cosmopolitan throughout Africa and India’s dry and semi-arid regions. Cumbu, black millet, spiked millet and candle millet are popular names while sajja is the local name in Andhra Pradesh. It is naturally cross pollinated (Allogamous) and the adaptation for cross pollination is Protogyny. Wind is the primary pollinator (anemophily).
Pearl millet is majorly grown in Rajasthan, Uttar Pradesh, Gujarat, Haryana, Karnataka, Maharashtra and Andhra Pradesh. In India, Pearl millet is cultivated in 7.65 million hectares with a production of 10.86 million tones and 1420 kg ha-1 productivity. In Andhra Pradesh, pearl millet is cultivated in 0.31 lakh hectares with a production of 0.71 lakh tones and a productivity of 2281 kg ha-1 (Directorate of Economics and Statistics, 2021).
Pearl millet is commonly known as a poor man’s food and important among nutritious cereals. The composition of grains (per 100 g) reveals carbohydrates (67.5 g), protein (11.6 g), fat (5 g), fiber (1.2 g), mineral matter (2.3 g), calcium (42 mg) and phosphorus (296 mg). The grain consists high amount of vitamins (thiamine, riboflavin and niacin) and minerals (P, K, Mg, Fe, Zn, Cu and Mn). It being rich source of energy is comparable to rice, wheat, maize and sorghum. Protein content of pearl millet is higher than barley (11.5%), maize (11.1%), sorghum (10.4%) and rice (7.2%). It has a low glycemic index (GI), which helps in weight loss and aids in cholesterol reduction (ICAR – AICRP on pearl millet, 2018).
For yield improvement in any crop, it is essential to develop genetically stable genotypes having high yield potential. It is therefore, necessary to estimate relative amounts of genetic and non-genetic variability exhibited by different characters using suitable parameters like genotypic coefficient of variation (GCV), phenotypic coefficient of variation (PCV) heritability (H) and genetic advance.
MATERIAL AND METHODS
In the present study 70 inbred lines of pearl millet were used. The experiment was conducted at Agricultural Research Station, Perumallapalle, Tirupati, during Rabi 2021. Experiment was laidout in randomized block design with two replications. In each replication every genotype was planted in three rows of three meters length by adopting 60 cm × 15 cm. All the recommended package of practices was adopted during entire crop season to raise healthy crop. Observations were recorded for 21 characters viz., days to 50% flowering, days to maturity, SPAD chlorophyll meter reading at 45 DAS, SPAD chlorophyll meter reading at 65 DAS, specific leaf area at 45 DAS (cm2 g-1), specific leaf area at 65 DAS (cm2 g-1), leaf sheath length (cm), leaf blade
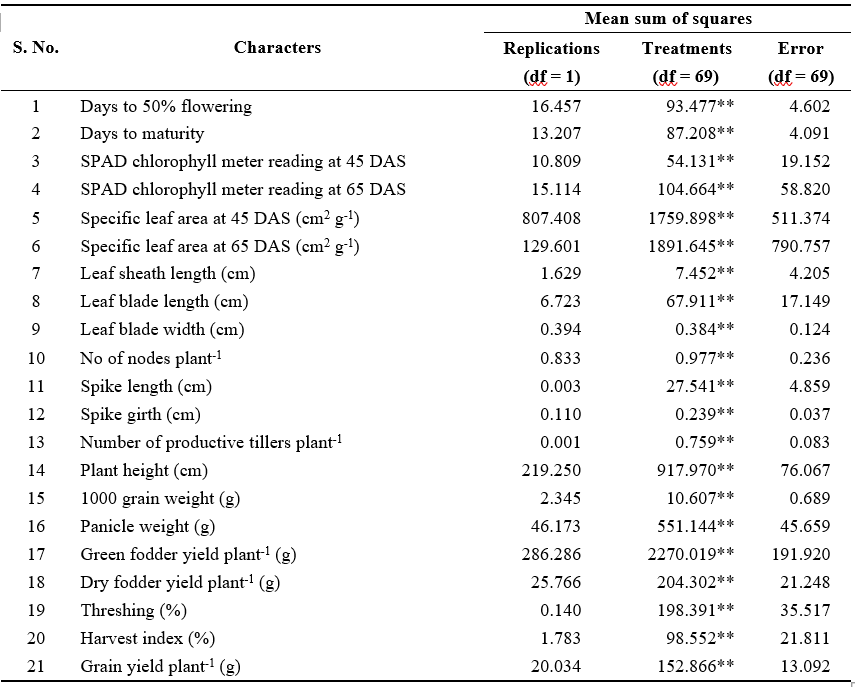
Table 1. Analysis of variance for yield and yield attributes in 70 inbred lines of pearl millet
length (cm), leaf blade width (cm), number of nodes plant-1, spike length (cm), spike girth (cm), number of productive tillers plant-1, plant height (cm), 1000 grain weight (g), panicle weight (g), green fodder yield plant-1 (g), dry fodder yield plant-1 (g), threshing (%), harvest index (%) and grain yield plant-1 (g). Data was subjected to statistical analysis for assessing phenotypic and genotypic coefficient of variation, heritability and genetic advance of percent of mean. Phenotypic and genotypic coefficients of variability were computed according to the methods suggested by Burton (1952). Heritability in broad sense was calculated as per the formula given by Allard (1960). Genetic advance was expressed as per cent of mean by using the formula suggested by Johnson et al. (1955).
RESULTS AND DISCUSSION
Analysis of variance for 70 pearl millet inbred lines revealed significant differences for all traits studied indicating the presence of considerable genetic variability among the genotypes studied (Table 1). Phenotypic coefficient of variance (PCV) was higher than genotypic coefficient of variance (GCV) for the traits studied indicating less interaction of traits with environment presented in Table 2.
The characters such as number of productive tillers plant-1 (GCV: 34.70%; PCV: 38.74%), 1000 grain weight (GCV: 21.75%; PCV: 23.22%), panicle weight (GCV: 40.89%; PCV: 44.43%), green fodder yield plant-1 (GCV: 39.52%; PCV: 43.01%), dry fodder yield
Table 2. Estimates of mean, range and genetic parameters for yield and yield attributes in 70 inbred lines of pearl millet
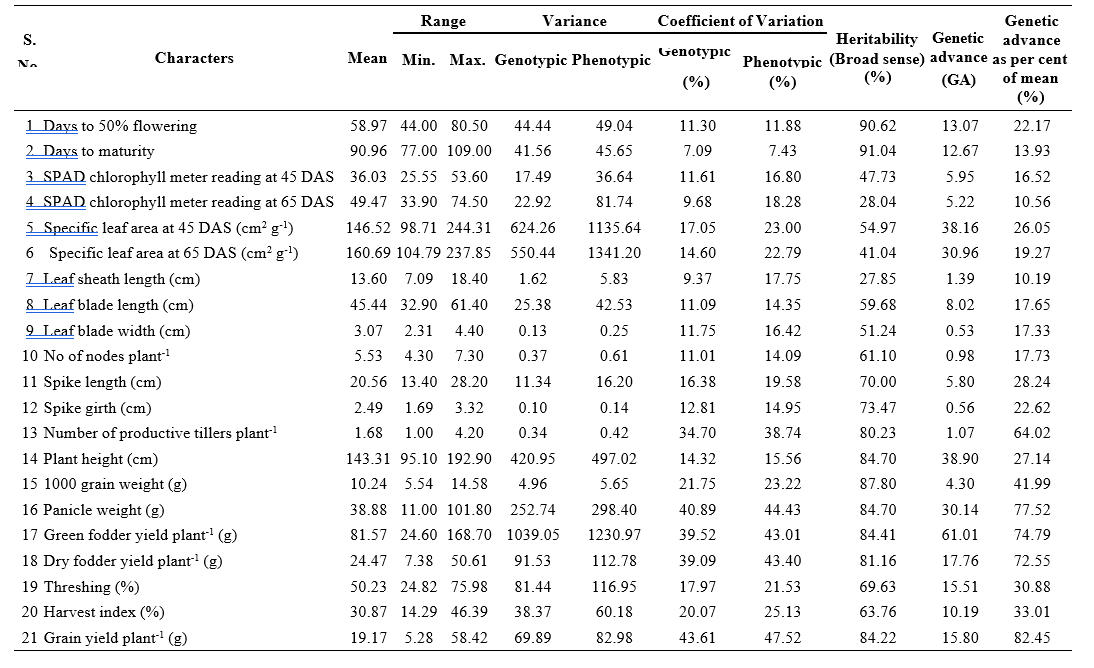
plant-1 (GCV: 39.09%; PCV: 43.40%) and harvest index (GCV: 20.07%; PCV: 25.13%) and grain yield plant-1 (GCV: 43.61%; PCV : 47.52%) showed higher estimates of coefficient of variation indicating the ample amount of variation among the inbred lines. Therefore, simple selection would be effective for further improvement of these characters.
Moderate estimates of coefficient of variation were observed for days to 50% flowering (GCV: 11.30%; PCV: 11.88%), SPAD chlorophyll meter reading at 45 DAS (GCV: 11.61%; PCV: 16.80%), specific leaf area at 45 DAS (GCV: 17.05%; PCV: 23.00%), specific leaf area at 65 DAS (GCV: 14.60%; PCV: 22.79%), leaf blade length (GCV: 11.09; PCV: 4.35%), leaf blade width (GCV: 11.75%; PCV: 16.42%), number of nodes plant-1 (GCV: 11.01%; PCV: 14.09%), spike length (GCV: 16.38%; PCV: 19.58%), spike girth (GCV: 12.81%; PCV: 14.95%), plant height (GCV: 14.32%;
PCV: 15.56%) and threshing percentage (GCV: 17.97%; PCV: 21.53%). This indicated the existence of sufficient variability for attempting selection to improve these traits in the genotypes studied.
Low estimates of GCV and moderate estimates of PCV were observed for SPAD chlorophyll meter reading at 65 DAS (GCV: 9.68%; PCV:18.28%) and leaf sheath length (GCV: 9.37%; PCV: 17.75%). The character days to maturity (GCV: 7.09%; PCV: 7.43%) exhibited low estimates of coefficient of variation.
Out of 21 traits studied, high heritability was observed for 14 characters viz., days to maturity (91.04%), days to 50% flowering (90.62%), 1000 grain weight (87.80%), plant height (84.70%), panicle weight (87.70%), green fodder yield plant-1 (84.41%), grain yield plant-1 (84.22%), dry fodder yield plant-1 (81.16%), number of productive tillers plant-1 (80.23%), spike girth (73.47%), spike length (70.00%), harvest index (63.76%), threshing percentage (69.63%) and number of nodes plant-1 (61.10%) in decreasing order of magnitude indicating that these characters were less influenced by environment. Higher values of heritability indicated that it may be due to higher contribution of genetic component and these traits were expected to remain stable under varied environmental conditions. Therefore, for improving these traits the selection will be more effective in early generation on the basis of per se performance of these traits.
Moderate estimates of heritability were observed for leaf blade length (59.68%), specific leaf area at 45 DAS (54.97%), leaf blade width (52.24%) SPAD chlorophyll meter reading at 45 DAS (47.73%) and specific leaf area at 65 DAS (41.04%).
Lower estimates of heritability were observed for SPAD chlorophyll meter reading at 65 DAS (28.04%), leaf sheath length (27.85%).
The characters viz., grain yield plant-1 (82.45%), panicle weight (77.52%), green fodder yield plant-1 (74.79%), dry fodder yield plant-1 (72.55%), number of productive tillers plant-1 (64.02%), 1000 grain weight (41.99%), harvest index (33.01%), threshing percentage
(30.88%), spike length (28.24%), plant height (27.14%), specific leaf area at 45 DAS (26.05%), spike girth (22.62%) and days to 50% flowering (22.17%) exhibited high genetic advance as per cent of mean. Moderate estimates of genetic advance as percent of mean were exhibited by specific leaf area at 65 DAS (19.27%), number of nodes plant-1 (17.73%), leaf blade length (17.65%), leaf blade width (17.33%), SPAD chlorophyll meter reading at 45 DAS (16.52%), days to maturity (13.93%), SPAD chlorophyll meter reading at 65 DAS (10.56%) and leaf sheath length (10.19%).
In the present investigation, high heritability coupled with high genetic advance as percent of mean was observed days to 50% flowering (h2 = 90.62%, GAM = 22.17%), spike length (h2 = 70.00%, GAM = 28.24%), spike girth (h2 = 73.47%, GAM = 22.62%), number of productive tillers plant-1 (h2= 80.23%, GAM = 64.02%), plant height (h2 = 84.70%, GAM = 27.14%), 1000 grain weight (h2 = 87.80%, GAM = 41.99%),panicle weight (h2b = 84.70%, GAM = 77.52%), green fodder yield plant-1 (h2 = 84.41%, GAM = 74.79%), dry fodder yield plant-1 (h2b = 81.16%, GAM = 72.55%), threshing percentage (h2b = 69.63%, GAM = 30.88%), harvest index (h2 = 63.76%, GAM = 33.01%) and grain yield plant-1 (h2 = 84.22%, GAM = 82.45%) indicating the predominance of additive gene action. . Early and simple selection could be exercised due to fixable additive gene effects.
High heritability coupled with moderate genetic advance as per cent of mean was recorded for days to maturity (h2= 91.04%, GAM = 13.93%) and number of nodes plant-1 (h2 = 61.10%, GAM = 17.73%) indicating that these characters were governed by additive gene effects and may express consistently in succeeding generations leading to greater efficiency of breeding programme.
Moderate heritability coupled with high genetic advance as per cent of mean was observed for specific leaf area at 45 DAS (h2= 54.97%, GAM = 26.05%), while SPAD chlorophyll meter reading at 45 DAS (h2 = 47.73%, GAM = 16.52%), specific leaf area at 65 DAS (h2= 41.04%, GAM = 19.27%), leaf blade length (h2 = 59.68%, GAM = 17.65%) and leaf blade width (h2= 51.24%, GAM = 17.33%) registered moderate estimates of both heritability and genetic advance as per cent of mean which indicated the preponderance of non-additive gene action. Hence, it could be suggested that improvement of these characters might be difficult through simple selection.
Low heritability coupled with moderate genetic advance as per cent of mean was observed for SPAD chlorophyll meter reading at 65 DAS (h2 = 28.04%, GAM = 10.56%) and leaf sheath length (h2 = 27.85%, GAM = 10.89%). It indicates that the character is highly influenced by environmental effects and selection would be ineffective.
To sum up with genetic parameters, high GCV, heritability and genetic advance as per cent of mean were observed for number of productive tillers plant-1, 1000 grain weight, panicle weight, green fodder yield plant-1, dry fodder yield plant-1, threshing percentage, harvest index and grain yield plant-1 indicating scope for genetic improvement through simple selection for all these traits with an implication of genetic variation mainly due to the presence of additive gene action.
ACKNOWLEDGEMENT
The authors would like to acknowledge Acharya N.G. Ranga Agricultural University, Lam, Guntur for granting financial assistance and support in the conduct of experiment at ARS, Perumallapalle, Tirupati, A.P. We gratefully thank the Professor and Head, Department of Genetics and Plant Breeding, S.V Agricultural college Tirupati for the suggestions made during conducting research work. This study is a part of M.Sc. student thesis work of first author.
LITERATURE CITED
Bhasker, K., Shashibushan, D., Krishna, K.M and Bhave, M.H.V. 2017. Genetic variability, heritability and genetic advance of grain yield in pearl millet [Pennisetum glaucum (L.) R. Br.]. International Journal of Pure and Applied Bioscience. 5(4): 1228-1231.
Bika, N.K and Shekawat, S.S. 2015. Genetic variability study in pearl millet [Pennisetum glaucum (L.) Br.] for green fodder yield and related traits. Electronic Journal of Plant Breeding. 6(2): 600-602.
Bind, H., Bharti, B., Kumar, S., Pandey, M.K., Kumar, D and Vishwakarma, D.N. 2015. Studies on genetic variability for fodder yield and its contributing characters in bajra [Pennisetum glaucum (L.) R. Br.]. Agricultural Science Digest. 35 (1): 78-80.
Burton, G.W. 1952. Quantitative inheritance in grasses. Proceedings of Sixth International Grassland Congress. 1: 277-283.
Dehinwal, A.K., Yadav, Y.P., Sivia, S.S and Kumar, A. 2016. Studies on genetic variability for different biometrical traits in pearl millet [Pennisetum glaucum (L.) R. Br.]. An International Quarterly Journal of Life Sciences. 11(2):1023-1026.
Directorate of Economics and Statistics, Department of Agriculture, cooperation and Farmers Welfare, Ministry of Agriculture and Farmers Welfare, GOI. 2021. Agricultural statistics at a glance. 56-58.
ICAR – All India Coordinated Research project on pearl millet, Jodhpur, Rajasthan. 2018. http://www. aicpmip.res.in.
Johnson, H.W., Robinson, H.F and Comstock, R.E. 1955. Estimation of genetic and environmental variability in soybean. Agronomy Journal. 47: 314-318.
Kaushik, J., Vart, D., Kumar, M., Nehra, M and Kumar, 2018. Genetic diversity assessment of pearl millet maintainer lines. Journal of Pharmocognosy and Phytochemistry. 7(5): 2428-2432.
Kumar, S., Babu, C., Revathi, S and Sumathi, P. 2017. Estimation of genetic variability, heritability and association of green fodder yield with contributing traits in fodder pearl millet [Pennisetum glaucum (L.) R. Br.]. International Journal of Advanced Biological Research. 7(1): 119-126.
Kumar, Y., Lamba, R.A.S., Yadav, H.P., Kumar, R and Vart, D. 2014. Studies on variability and character association under rainfed conditions in pearl millet [Pennisetum glaucum (L.) R. Br.] hybrids. Forage research. 39(4): 175-178.
Lush, J.L. 1940. Intra-sire correlation and regression of offspring in ramsasa method of estimating heritability of characters. Proceedings of American Society of Animal Production. 33: 292-301.
Narasimhulu, R., Reddy, B.S., Satyavathi, C.T and Ajay, B.C. 2021. Performance, Genetic Variability and Association Analysis of Pearl Millet Yield Attributing Traits in Andhra Pradesh’s Arid Region. Chemical Science Review and Letters. 10 (38): 177-182.
Nehra, M., Kumar, M., Kaushik, J., Vart, D., Sharma, R.K and Punia, M.S. 2017. Genetic divergence, character association and path coefficient analysis for yield attributing traits in pearl millet [Pennisetum glaucum (L.) R. Br] inbreds. Chemical Science Review and Letters. 6(21): 538-543.
- Genetic Divergence Studies for Yield and Its Component Traits in Groundnut (Arachis Hypogaea L.)
- Correlation and Path Coefficient Analysis Among Early Clones Of Sugarcane (Saccharum Spp.)
- Character Association and Path Coefficient Analysis in Tomato (Solanum Lycopersicum L.)
- Survey on the Incidence of Sesame Leafhopper and Phyllody in Major Growing Districts of Southern Zone of Andhra Pradesh, India
- Effect of Organic Manures, Chemical and Biofertilizers on Potassium Use Efficiency in Groundnut
- A Study on Growth Pattern of Red Chilli in India and Andhra Pradesh

