Genetic Divergence Studies for Yield and Its Component Traits in Groundnut (Arachis Hypogaea L.)
0 Views
P. SUSMITHA*, A. PRASANNA RAJESH, M. REDDI SEKHAR AND SK. NAFEEZ UMAR
Department of Genetics and Plant Breeding, S.V. Agricultural College, ANGRAU, Tirupati-517 502.
ABSTRACT
Mahalanobis D2 statistic was used to quantify the genetic diversity among 30 advanced breeding lines of groundnut for
fifteen traits. Based on Tocher’s method of clustering, the genotypes were grouped into five clusters. Among these five clusters, Cluster I had maximum number of 26 genotypes and remaining four clusters i.e., II, III, IV and V were solitary clusters with only one genotype each. The maximum inter- cluster distance was observed between cluster III and V followed by cluster II and V, cluster II and IV and cluster I and V.Considering the cluster distances and cluster means, cross combinations of clusters III X V and II X V is advised in order to obtain transgressive segregants for yield and yield parameters.Further, kernel yieldperplantwas observed to contribute maximum, followed by pod yieldper plant towards the total divergence of the genotypes studied in the present investigation.
KEYWORDS: Groundnut, D2 and genetic divergence.
INTRODUCTION
Groundnut(Arachis hypogaea L.) is an allotetraploid, self-pollinating member of the Fabaceae family with chromosome number 2n = 4x = 40. It is a major oilseed crop and grain legume that is grown annually in tropical and subtropical regions of the world. It is indigenous to South America and goes by a variety of regional names, including wonder legume, peanut, earthnut, goober peas, monkey nut, and manila nut. Globally, India is the second largest producer with 101 lakh tonnes and with productivity of 1863 kg ha-1 in 2021-22 (agricoop.nic.in). It is the world’s third-most important source of vegetable protein and the fourth-most significant source of edible oil. In Andhra Pradesh, groundnut covers an area of 5.94 lakh ha with production of 6 lakh tonnes and productivity of 1011 kg ha-1, respectively (Directorate of Economics and Statistics, Govt. of A.P, India, 2022-2023).
The information on genetic diversity in a crop is essential in order to have breeding programmes for higher yield potentials. Success of any plant breeding programme largely depends on the choice of appropriate parents. Utilization of genetically divergent parents in hybridization results in expansion of genetic base and development of superior recombinants. The genetic divergence analysis determines the variation pattern in the collection of genotypes, identifies the characters, and differentiates the genotypes into different groups that could be used in the crop improvement program.
In light of this, the current study used the Mahalanobis D2 statistic to categorise and comprehend the nature and extent of genetic diversity among the groundnut genotypes of the Agricultural Research Station, Kadiri of Andhra Pradesh.
MATERIAL AND METHODS
The experimental seed material for the present investigation comprised of 30 diverse groundnut advanced breeding lines. The seed material for the study was obtained from Agricultural Research Station, Kadiri of ANGRAU.
These genotypes were sown during Rabi 2023 in a Randomized Block Design (RBD) with three replications. In each replication, every genotype was sown in continuous three rows plots of 3 m length with a spacing of 30 cm between the rows and 10 cm between the plants within the row. The crop was raised under irrigated conditions and all recommended practices were followed to raise a healthy crop. Observations were recorded on yield and yield attributing traits namely, days to first flowering, days to 50 per cent flowering, days to maturity, plant height, number of mature pods per plant, number of immature pods per plant, dry haulm yield per plant (g), pod yield per plant (g), kernel yield per plant (g), shelling per cent, hundred kernel weight (g), sound mature kernel (per cent), harvest index (per cent), oil content (per cent) and protein content (per cent).Observations were recorded for all the genotypes separately on randomly chosen five competitive plants in each genotype in each replication for all the characters except for days to first flowering, days to 50 per cent flowering and days to maturity, which were recorded on plot basis.
The data collected was subjected to standard statistical analysis. Genetic diversity in the material was analyzed using Mahalanobis D2 statistic, 1936 and expanded by Rao, 1952 and the genotypes were grouped into different clusters according to Tocher’ method. In order to determine the intra- and inter-cluster distances, the Singh andChaudhary’s approach, 1985 was employed. The divergence contribution of each character was computed using Singh’s, 1981 approach.
RESULTS AND DISCUSSIONS
Based on D2 value, thirty groundnut genotypes were grouped into five clusters (Table 1). Among these clusters, cluster I was the largest group comprising of 26 genotypes and remaining all four clusters i.e., II, III, IV and V contain one genotype each.
Inter-cluster distance (Table 2) was considerably larger than intracluster distance which indicated the presence greater diversity among the genotypes of distant group. These findings were in conformity with the previous reports of Zaman et al. (2010), Raghuwanshi et al. (2016), Saini et al. (2020), Kulheri et al. (2023), Sai Teja et al. (2023).
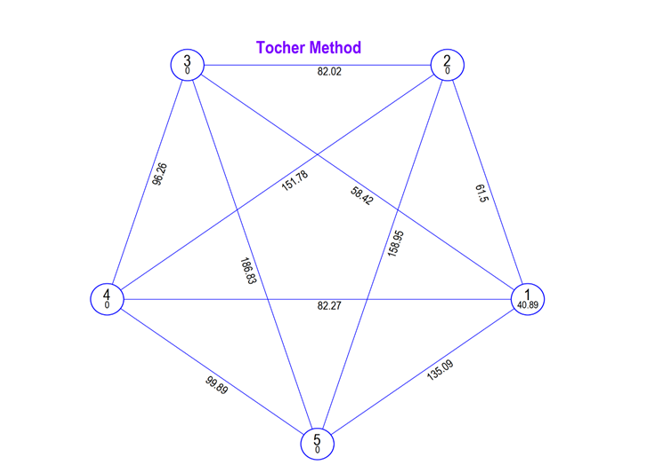
Fig. 1. Tocher’s diagram depicting inter and intra cluster (D2) distances among five clusters in advanced breeding lines of groundnut.
The inter-cluster distance analysis revealed that cluster III and V had the highest divergence (186.83), followed by cluster II and V (158.95), cluster II and IV (151.78) and cluster I and V (135.09). The genotypes in these clusters can be utilised as parents for hybridization because they showed comparatively high divergence when compared to other clusters. Whereas, minimum inter- cluster distance was identified between cluster I and III (58.42) and cluster I and cluster II (61.50). The only intra cluster distance was observed in cluster I (40.49), While other clusters were mono-genotypic with no intra-cluster divergence.
Cluster mean values for various characters (Table 3) indicated that cluster IV recorded highest cluster mean for dry haulm yield per plant, kernel yield per plant and 100 kernel weight and cluster V exhibited highest cluster mean for number of mature pods per plant, pod yield per plant and harvest index. Cluster III was the best source for days to first flowering, days to maturity, number of immature pods per plant, shelling per cent and protein content, while cluster II performed best for days to 50 per cent flowering, plant height, sound mature kernel and oil content indicating the importance of selection of genotypes from the corresponding clusters in hybridization programs for effective improvement of the respective traits.
Contribution of various characteristics to total divergence (Table 4) revealed that kernel yield per plant contributed the most to the diversity of all the characters, taking 63 times out of 435 combinations and contributed maximum towards total divergence (14.53%) followed by pod yield per plant (12.54%), dry haulm yield per plant (9.42%), shelling per cent (8.8%), plant height (8.74%), sound mature kernel (8.32%), harvest index (7.36%), number of mature pods per plant (6.87%) , hundred kernel weight (6.21%), number of immature pods per plant (5.23%), days to 50% flowering (3.43%), days to first flowering (2.56%), protein content (2.46%), days to maturity (2.3%) and oil content (1.23%).It has been suggested that the character with maximum contribution towards divergence should be prioritized for future hybridization projects. The greatest contribution of kernel yield per plant towards divergence was also earlier reported by Dudhatra et al. (2022).
LITERATURE CITED
agricoop.nic.in. 2021-22 https://agriwelfare.gov.in/Documents/ CWWGDATA/Agricultural_Statistics_at_a_ Glance_2022_0.pdf
Directorate of Economics and Statistics, Govt. of A.P, India, 2022-2023. https://des.ap.gov.in/jsp/social/ SEASONANDCROPREPORT_2022_23.pdf.
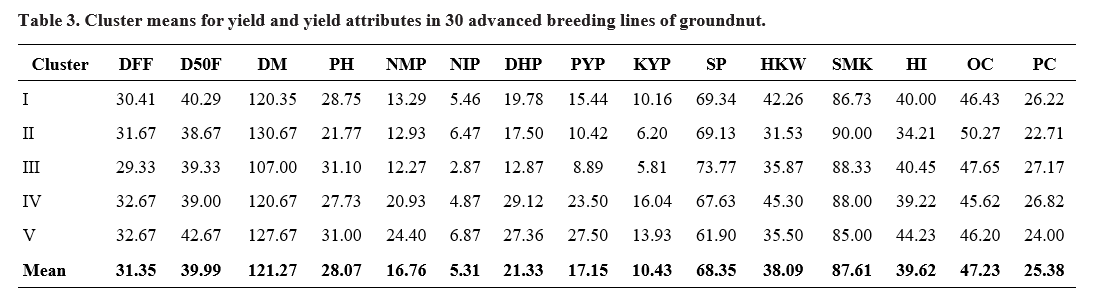
DFF: Days to first flowering; D50F: Days to 50% flowering; DM: Days to maturity; PH: Plant height (cm); NMP: No. of mature pods plant-1; NIP: No. of immature pods plant-1; DHP: Dry haulm yield plant-1 (g); PYP: Pod yield plant-1 (g); KYP: Kernel yield plant-1 (g); SP: Shelling per cent; HKW: Hundred kernel weight (g); SMK: Sound mature kernel (%); HI: Harvest index (%); OC: Oil content (%); PC: Protein content (%)
Dudhatra, R.S., Viradiya, Y.A., Joshi, K.B., Desai, T.A and Vaghela, G.K. 2022. Genetic divergence analysis in groundnut (Arachis hypogaea L.) genotypes. Emergent Life Sciences Research. 8(1): 114-118.
Kulheri, A., Sikarwar, R.S., Jakhar, K and Rajput, S.S. 2023. Genetic Divergence Study in Groundnut Genotypes (Arachis hypogaea L.). International Journal of Environment and Climate Change. 13(9): 2512-2517.
Mahalonobis, P.C. 1936. On the generalized distance in statistics. Proceedings of the National Institute of Science of India. 12: 49-55.
Raghuwanshi, S.S., Kachhadia, V.H., Vachhani, J.H., Jivani, L.L and Patel, M.B. 2016. Genetic divergence in groundnut (Arachis hypogaea L.). Electronic Journal of Plant Breeding. 7(1): 145-151.
Rao, C.R. 1952. Advanced Statistical Methods in Biometrical Research. John Wiley and sons. New York. 357-363.
Saini, H., Sharma, M.M., Saini, L.K., Solanki, K.L and Singh, A.G. 2020. Genetic divergence analysis in groundnut (Arachis hypogaea L.). International Journal of Chemical Studies. 8(1): 1781-1784.
Saiteja, R., John, K., Devi, M.S and Murthy, B.R. 2023. Studies on genetic divergence for yield, yield components and quality traits in groundnut (Arachis hypogaea L.). Andhra Pradesh Journal of Agricultural Sciences. 9(3): 205-209.
Singh, D. 1981. The relative importance of characters affecting divergence. Indian Journal Genetics. 41: 237-245.
Singh, R.K and Chaudhary, B.D. 1977. Biometrical Methods in Quantitative Genetic Analysis. Kalyani Publishers, New Delhi. 215-218.
Zaman, M.A., Tuhina-Khatun, M., Moniruzzaman, M and Yousuf, M.N. 2010. Genetic divergence in groundnut (Arachis hypogaea L.). Bangladesh Journal of Plant Breeding and Genetics. 23(1): 45-49.
- Effect of Sowing Window on Nodulation, Yield and Post – Harvest Soil Nutrient Status Under Varied Crop Geometries in Short Duration Pigeonpea (Cajanus Cajan L.)
- Nanotechnology and Its Role in Seed Technology
- Challenges Faced by Agri Startups in Andhra Pradesh
- Constraints of Chcs as Perceived by Farmers in Kurnool District of Andhra Pradesh
- Growth, Yield Attributes and Yield of Fingermillet (Eleusine Coracana L. Gaertn.) as Influenced by Different Levels of Fertilizers and Liquid Biofertilizers
- Consumers’ Buying Behaviour Towards Organic Foods in Retail Outlets of Ananthapuramu City, Andhra Pradesh


