Evaluation of Groundnut (Arachis Hypogaea L.) Genotypes for Yield, Yield Attributes and Water Use Efficiency Related Traits
0 Views
S. DIVYA SREE*, B. RUPESH KUMAR REDDY, M. SHANTHI PRIYA AND A.R. NIRMAL KUMAR
Department of Genetics and Plant Breeding, S.V. Agricultural College, ANGRAU, Tirupati-517502.
ABSTRACT
Thirty-six groundnut genotypes were grown during Rabi, 2021-2022 in order to evaluate the performance of elite geno- types of groundnut for 14 characters. The experiment was laid out in randomized block design with three replications. Analysis of variance showed that mean sum of squares due to genotypes was highly significant for all characters studied. For pod yield plant-1; the genotypes TCGS-2223, TCGS-2040, TCGS-2039, TCGS-2230, TCGS-2053, TCGS-2055, TCGS-2317, TCGS-2278,
TCGS-2233 and TCGS-2052 showned improved performance. TCGS-2223 exhibited better performance for all the characters studied except for the trait plant height. TCGS-2053 outperformed all the other genotypes in terms of pod yield plant-1.
KEYWORDS: Genotypes, RBD, Pod yield, Plant height.
INTRODUCTION
Cultivated groundnut or peanut (Arachis hypogaea L.) is an annual self pollinated legume crop with chromosome number of 2n = 4x = 40. It is the only legume crop in which pods are formed inside the soil. It belongs to the family fabaceae. It is native to South America, grown throughout the tropical and sub-tropical regions of the world between the latitudes of 40o N to 40o S.
It is an important oilseed crop known for its several purposes including edible oil production, direct human consumption as food and for animal consumption in the form of silage, hay and oilcake. It is a rich source of high quality oil (44-56%), protein (22-30%) on dry seed basis, carbohydrates (10-25%), vitamins (E and B complex), minerals (Ca, P, Mg, Zn and Fe) and fiber. Being a legume it adds nitrogen and organic matter to soil.
It is grown as a sole crop, intercrop or mixed crop (Rao et al. 1990; Ghosh et al. 2004). With its multiple uses; groundnut is now a great cash crop for both local and international trade in a number of emerging and developed nations.
Globally, it is cultivated in an area of 29.92 Mha with annual production of 55.30 Mt and productivity of 1851 kg ha-1 (FAOSTAT, 2021). In India, groundnut covers an area of 6.09 Mha with a production of 10.21 Mt and productivity of 1676 kg ha-1. In Andhra Pradesh, it is cultivated in an area of 0.87 Mha with a production of 0.78 Mt and productivity of 894 kg ha-1 (Directorate of Economics and Statistics, 2021).
Mainly groundnut is cultivated as a rainfed crop but is also grown as irrigated crop. During rabi season, due to the depletion of ground water resources, water shortage is usual during the crop period. Hence, there is a need to find out Water Use Efficiency (WUE) genotypes in groundnut. WUE is an important trait which contributes to productivity when there are limited water resources. The productivity of the crop can be enhanced by identifying the genotypes that utilise the scarce resources of water more efficiently (Arunkumar et al., 2017).
WUE in groundnut is correlated with the specific leaf area (SLA) and SPAD Chlorophyll Meter Reading (SCMR). SCMR and SLA can be utilised as surrogate traits to choose genotypes with high WUE (Sab et al. 2018). These physiological traits will greatly aid us in bringing genetic improvements for WUE in groundnut genotype improvement ultimately leading to the evolution of better genotypes adapted to drought conditions, when used as selection criteria in breeding programmes or in the selection of efficient genotypes.
Considering the above factors, the research was undertaken to study the per-se performance for yield, yield attributes and water use efficiency related traits in 36 genotypes of groundnut.
MATERIAL AND METHODS
The experiment was laid out in Randomized Block Design (RBD) with three replications and 36 genotypes of groundnut were evaluated at dry land farm of S.V. Agricultural College, Tirupati, during rabi, 2021-2022. The data was recorded on fourteen traits viz., days to 50%
flowering, days to maturity, plant height (cm), number of primary branches plant-1, number of secondary branches plant-1, kernel yield plant-1 (g), hundred kernel weight (g), shelling per cent, sound mature kernel per cent, harvest index (%), SPAD Chlorophyll Meter Reading at 60 DAS, Specific Leaf Area at 60 DAS (cm2 gm-1), Relative Water Content (%) and pod yield plant-1 (g). The data was analyzed through software – INDOSTAT.
RESULTS AND DISCUSSION
The phenotypic variation manifested by the genotype has two components namely genotypic and environmental. Analysis of variance among 14 characters revealed significant differences among the means of all the genotypes. Thus, it indicated considerable scope for selection of high yielding and genotypes with more yield and water use efficiency. The differences can be attributed to variations present in the genotypes as well as the environmental factors.
Mean performance of genotypes for different yield, yield attributes and water use efficiency related traits
-
Days to 50 % flowering
The mean number of days to 50% flowering ranged from 22.00 days (TCGS-2233) to 31.33 days (TCGS-2044). When compared to the average, nineteen genotypes reached blooming early (26.60 days). As a result, these genotypes can be used as donor parents in the hybridization programme to develop short duration genotypes.
2. Days to maturity
The mean number of days to maturity ranged from 101.67 (TCGS-2233) to 112.00 days (TCGS-2052).
Twenty one genotypes reached maturity sooner than the average of 106.25 days. In order to develop short duration genotypes, these genotypes can be used in the hybridization programme as donor parents.
3. Plant height (cm)
With a general mean of 35.41 cm, the mean values for plant height ranged from 25.67 cm (TCGS-2051) to 45.97 cm (TCGS-2052). Eighteen genotypes showed better per se performance than the average (35.41 cm).
4. Number of primary branches plant-1
Number of primary branches plant-1 count ranged from 3.87 (TCGS-2230) to 8.82 (TCGS-2053). Fifteen genotypes recorded more number of primary branches plant-1 than the average of 5.26.
5. Number of secondary branches plant-1
Number of secondary branches plant-1 were between 0.20 (TCGS-2068 and Narayani) and 2.73 (TCGS-2229). Nineteen genotypes recorded more secondary branches plant-1 than the average of 1.50.
6. Kernel yield plant-1 (g)
Kernel yield plant-1 mean varied from 7.74 g (Greeshma) to 15.36 g TCGS-2053 and twenty genotypes showed higher yields than general mean of 10.93 g.
7. Hundred kernel weight (g)
The mean values for hundred kernel weight ranged from 40.67 g (TCGS-2068) to 66.40 g. (TCGS-1798). Sixteen genotypes recorded hundred kernel weights greater than the average of 50.40 g.
8. Shelling per cent
Nineteen genotypes had higher shelling per cent than the overall mean of 71.73%, with mean values for shelling per cent ranging from 57.26% (TCGS-2230) to 79.87% (TCGS-2235).
9. Sound mature kernel (%)
With an overall mean value of 51.00%, the mean values for this character ranged from 34.03% (Tirupati-4) to 72.17% (Rohini). Seventeen genotypes registered sound mature kernel percentages that were higher than the average of 51.00%.
10. Harvest index (%)
Nineteen genotypes recorded higher harvest indices than the average of 51.17%. The genotype means varied from 24.95% (TCGS-2044) to 71.45% (TCGS-2053).
11. SPAD chlorophyll meter reading (SCMR) at 60 DAS
With a general mean of 47.55, the mean values for the SPAD chlorophyll meter reading (SCMR) at 60 DAS ranged from 43.30 (K-6) to 50.77 (TCGS-2223).
Eighteen genotypes obtained higher SPAD chlorophyll readings than the average of 47.55.
12. Specific leaf area at 60 DAS (SLA) (cm2 g-1)
The mean values of specific leaf area at 60 DAS from 119.22 cm2 g-1 (TCGS-1798) to 265.35 cm2 g-1 (TCGS-2068) and seventeen genotypes showed lower specific leaf area than their overall mean of 207.55 cm2 g-1.
13. Relative water content (%)
The mean percentages of relative water content ranged from 69.24% (TCGS-2060) to 92.53% (TCGS-
Table 1. Analysis of variance for yield, yield attributes and water use efficiency (WUE) related traits in 36 genotypes of groundnut
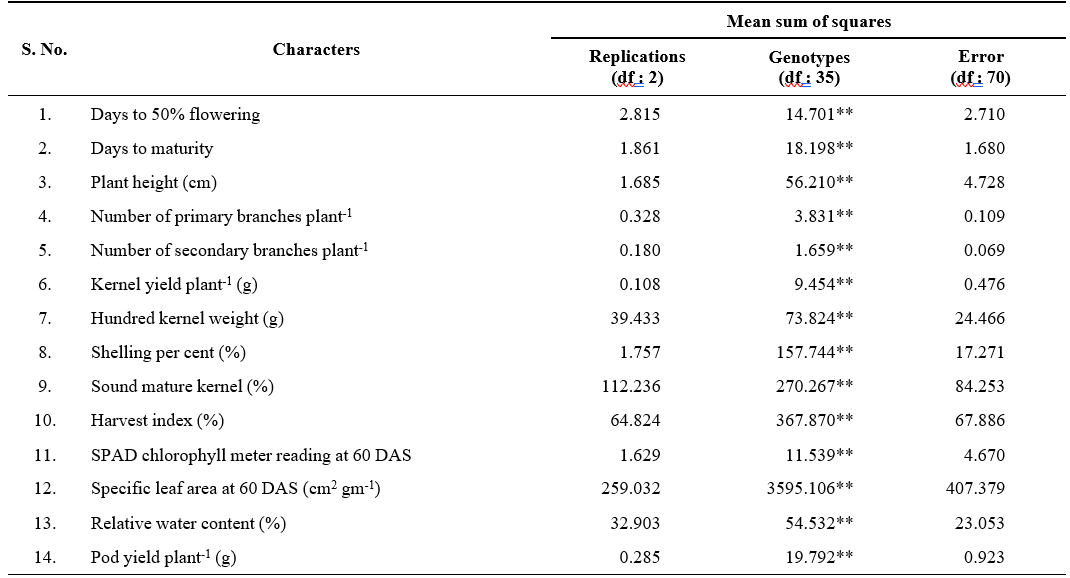
Table 2. Per se performance for yield, yield attributes and water use efficiency (WUE) related traits in 36 genotypes of groundnut
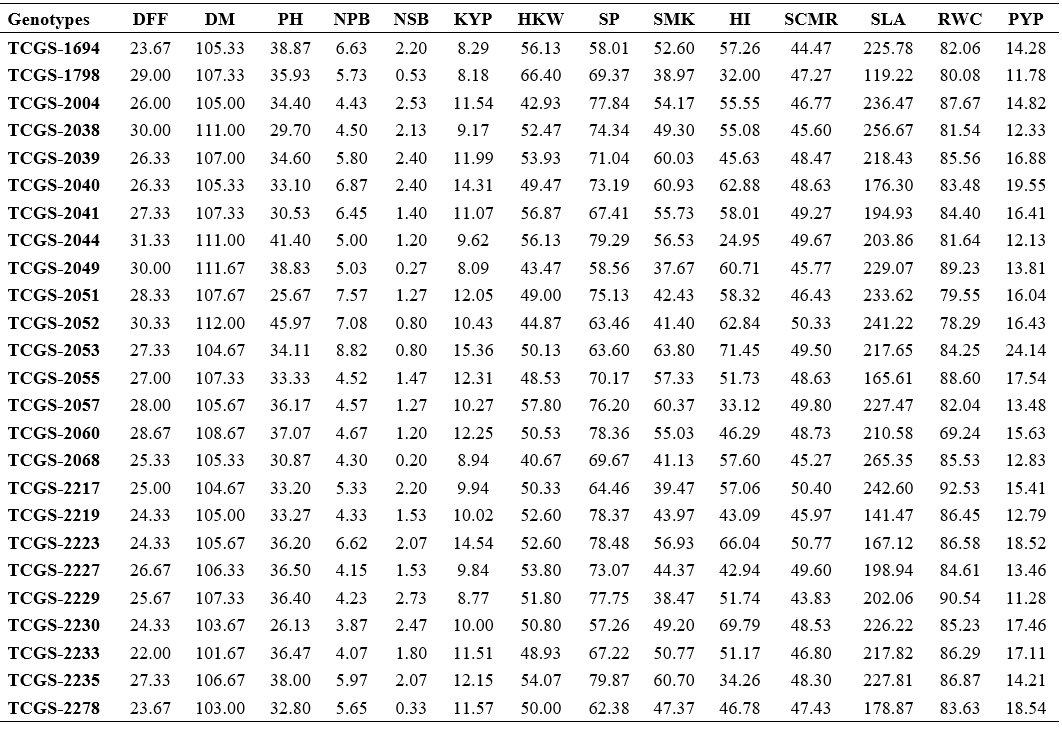
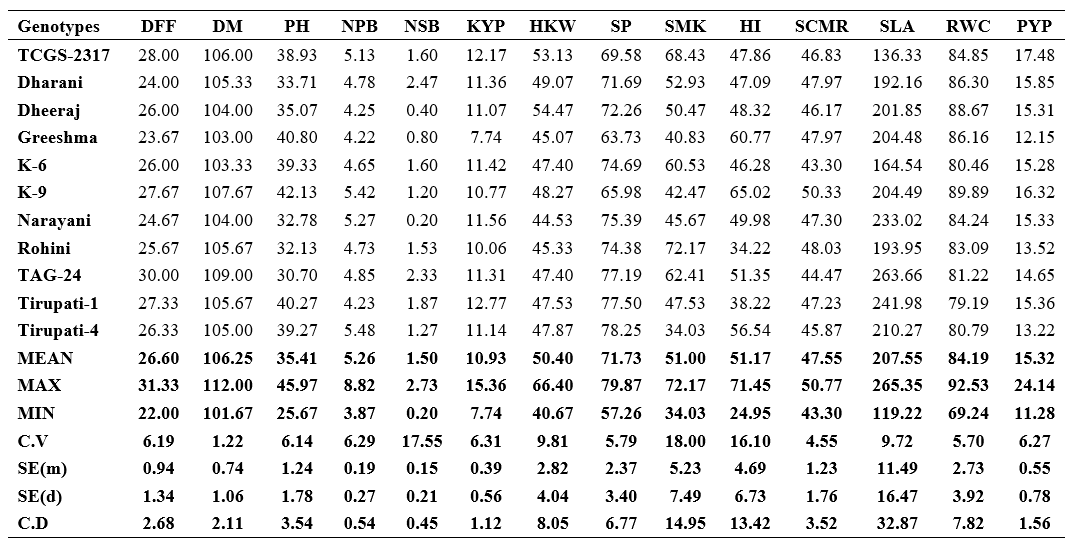
DFF : Days to 50% flowering; DM : Days to maturity; PH : Plant height (cm); NPB : Number of primary branches plant-1; NSB : Number of secondary branches plant-1; KYP : Kernel yield plant-1 (g); HKW : Hundred kernel weight (g); SP : Shelling per cent (%); SMK : Sound mature kernel (%); HI : Harvest index (%); SCMR : SPAD chlorophyll meter reading at 60 DAS; SLA : Specific leaf area at 60 DAS (cm2 gm-1); RWC : Relative water content (%); PYP : Pod yield plant-1 (g)
Table 3. List of top five genotypes based on per se performance in groundnut
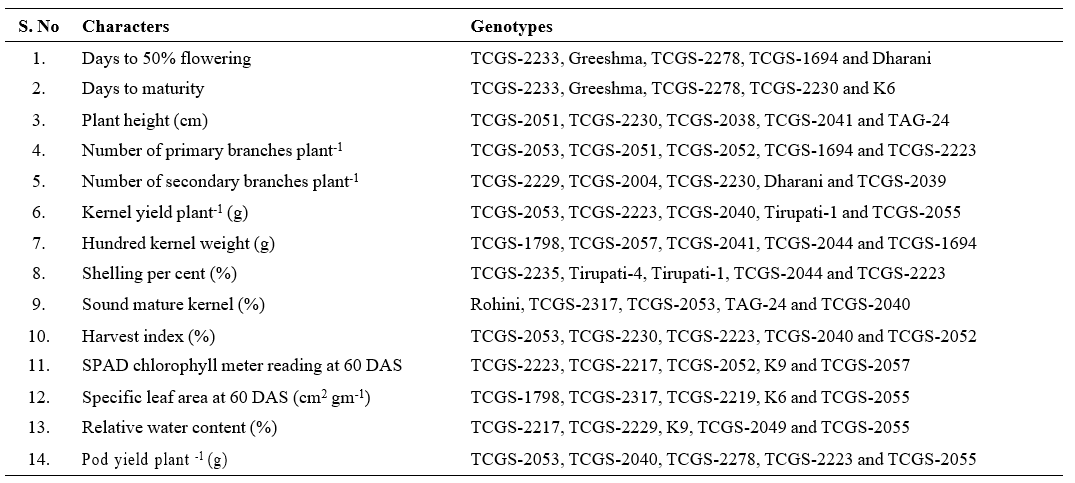
Table 4. Identification of promising genotypes for yield, yield attributes and water use efficiency (WUE) related traits in groundnut

DFF : Days to 50% flowering; DM : Days to maturity; PH : Plant height (cm); NPB : Number of primary branches plant-1; NSB : Number of secondary branches plant-1; KYP : Kernel yield plant-1 (g); HKW : Hundred kernel weight (g); SP : Shelling per cent (%); SMK : Sound mature kernel (%); HI : Harvest index (%); SCMR : SPAD chlorophyll meter reading at 60 DAS; SLA : Specific leaf area at 60 DAS (cm2 gm-1); RWC : Relative water content (%); PYP : Pod yield plant-1 (g)
2217). 21 genotypes revealed higher relative water content than their average (84.19%).
14. Pod yield plant-1 (g)
Genotypes for pod yield plant-1 ranged from 11.28 g (TCGS-2229) to 24.14 g (TCGS-2053) and
eighteen genotypes showed higher pod yield plant-1 than the general mean of 15.32 g.
The top five genotypes for each character was determined based on per se performance, and they were listed in Table 3. Table 4 provides a list of the promising genotypes that were identified for several traits. For pod yield plant-1; the genotypes TCGS-2223, TCGS-2040, TCGS-2039, TCGS-2230, TCGS-2053, TCGS-2055, TCGS-2317, TCGS-2278, TCGS-2233 and TCGS-2052
shown improved performance. These genotypes are deserving of use in the improvement of characters. Among the 36 genotypes examined, TCGS-2223 and TCGS- 2040 are adjudged as promising genotypes not only for high yield and yield components, but also for water use efficiency related traits such as SPAD chlorophyll meter readings, specific leaf area and relative water content. Therefore, these genotypes may be involved in the crossing programme for by duly estimating their general and specific combining abilities in the future breeding programmes. Further, multi-location testing of these entries over years could be recommended to test their suitability and stability.
LITERATURE CITED
Arunkumar, P., Maragatham, N., Panneerselvam, S., Ramanathan, S and Jeyakumar, P. 2017. Water requirement of groundnut under different intercropping system and WUE in groundnut equivalent rate. The Pharma Innovation Journal. 6(11): 322-325.
Directorate of Economics and Statistics, Department of Agriculture, cooperation and Farmers Welfare, GOI. 2021. Agricultural Statistics at a glance. 50- 51.
FAOSTAT, 2021. Food and Agriculture Organization of the United Nations. World Agricultural Production, Rome, Italy. http://faostat.fao.org/.
Ghosh, P.K. 2004. Growth, yield, competition and economics of groundnut/cereal fodder intercropping systems in the semi-arid tropics of India. Field Crops Research. 88(2-3), : 227-237.
Rao, R.C.N., Wadia, K.R and Williams, J.H. 1990. Intercropping short and long duration groundnut (Arachis hypogaea) genotypes to increase productivity in environments prone to end-of- season droughts. Experimental Agriculture. 26(1): 63-72.
Sab, S., Shanthala, J., Savithramma, D.L and Bhavya,
M.R. 2018. Study of genetic variability and character association for water use efficiency (WUE) and yield related traits advance breeding lines of groundnut (Arachis hypogaea L.). International Journal of Current Microbiology and Applied Sciences. 7(6): 3149-3157.
- Genetic Diversity Analysis of 64 Maize Inbred Lines for Yield Traits Using D2 Statistics and Principal Component Analysis
- Effect of Border Crops on Activity of Predatory Fauna In Blackgram (Vigna Mungo L.)
- Effect of Organic Nutrient Management Practices on Growth and Yield of Foxtail Millet
- Species Diversity of Sugarcane Shootborers in Major Sugarcane Growing Districts of Andhra Pradesh
- Faunistic Studies on Economically Important Lepidopterans in Different Field Crops of Tirupati District
- Production Potential of Sweet Corn as Influenced by Organic Manures and Foliar Nutrition

