Estimates of Variability, Heritability and Genetic Advance in Browntop Millet
0 Views
ADURU HITHAISHY*, M. SREEVALLI DEVI, B. SANTHOSH KUMAR NAIK, T.M. HEMALATHA AND M. REDDI SEKHAR
Department of Genetics and Plant Breeding, S.V. Agricultural College, ANGRAU, Tirupati-517 502.
ABSTRACT
The present study aimed to assess the genetic variability, heritability, and genetic advance for nine yield and yield- attributing traits in 30 genotypes of brown top millet (Brachiaria ramosa (L.) Stapf.) maintained at Agricultural Research Station, Perumallapalle during Rabi 2024-25. Significant differences among genotypes were observed for all traits, indicating substantial genetic variability in the material studied. High genotypic and phenotypic coefficients of variation were recorded for number of productive tillers per plant and grain yield per plant, suggesting ample scope for improvement through selection. Traits such as test weight, number of productive tillers per plant, harvest index and grain yield per plant exhibited high heritability coupled with high genetic advance as a percentage of mean, indicating the predominance of additive gene action and the effectiveness of phenotypic selection for these traits. The findings highlight productive tillers per plant, grain yield, harvest index and test weight as ideal targets for genetic improvement in brown top millet breeding programmes. Integrating variability measures with heritability and genetic gain estimates is emphasized for formulating effective breeding strategies.
KEYWORDS: Brown top Millet, GCV, PCV, Heritability and Genetic Advance.
INTRODUCTION
Bracharia ramosa (L.) Stapf. or Urochola ramosa (L.) is a minor millet originated from Southeast Asia (Sheahan, 2014). It was first domesticated in southern India and belongs to the family Poaceae and sub-family Panicoideae. Brown top millet is a self-pollinated crop with a chromosome number of 2n=4x=32. It is locally known as “Andukora” or pedda sama in Telugu and korlle or karlaki in Kannada. The crop is commonly called browntop millet because of its dark brown tinge on its seeds. It is also known as signal grass. Taxonomically, it is represented by var. ramosa and var. pubescens (Basappa et al.,1987).
Genetic variability is a key prerequisite for genetic improvement in plant breeding. Knowledge on the extent of variability existing in a crop species for different traits is crucial, because it serves as the basis for effective selection. To obtain a realistic indication of genetic variation in any trait, phenotypic variability must be partitioned into heritable and non-heritable components. The potential genetic gain from a selection process may be evaluated using estimations of heritability along with genetic advance. The aim of this research was to estimate the degree of variation among different yield and yield attributing traits and identify the best suitable lines for increasing the grain yield of brown top millet.
MATERIALS AND METHODS
The current study was carried out at Agricultural Research Station, Perumallapalle during rabi, 2024-25. The experimental material constituted 30 brown top millet genotypes and the experiment was conducted in Randomized Block Design with three replications and every entry was sown in three rows with a spacing of 45 cm x 10 cm. Timely management of recommended package was done during the crop period. Observations were noted for plant height, the number of productive tillers per plant, days to 50% flowering, panicle length, days to maturity, test weight, fodder yield per plant, grain yield per plant and harvest index.
RESULTS AND DISCUSSIONS
Analysis of variance was carried out for nine yield and yield attributing traits viz., plant height, number of productive tillers per plant, days to 50% flowering, panicle length, days to maturity, fodder yield, test weight, grain yield and harvest index is represented in Table.1 The results revealed significant differences among the studied genotypes for all the traits which indicate the
Table 1. Analysis of variance for yield, yield components in 30 browntop millet
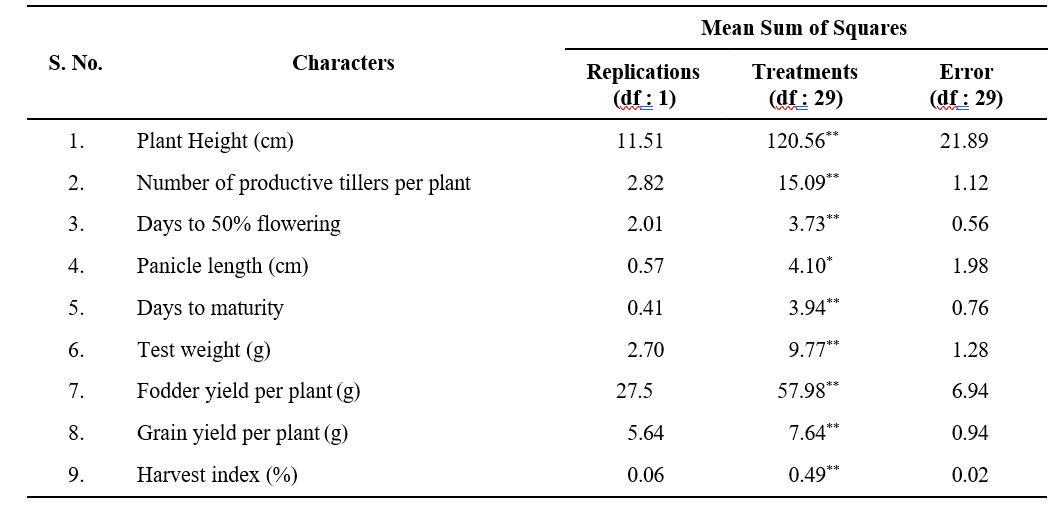
presence of substantial amount of genetic variability in the material investigated.
High GCV was recorded for number of productive tillers per plant (27.66) followed by grain yield per plant (25.37). Moderate GCV was recorded for harvest index (19.54) followed by test weight (13.87) and fodder yield per plant (10.06) and remaining triats showed low values (Table-2).
High PCV was recorded for number of productive tillers per plant (29.80) followed by grain yield per plant (28.71) and harvest index (22.04). Moderate PCV was recorded for test weight (14.42) followed by panicle length (13.74), fodder yield per plant (11.48) and Plant height (11.06).
High GCV and PCV was recorded for traits like grain yield per plant (GCV: 25.37 %; PCV: 28.71 %) and number of productive tiller per plant (GCV: 27.66%; PCV: 29.80 indicating the presence of ample amount of variation among the genotypes. Thus, simple selection would be effective for further improvement of these traits. These results are in accordance with Rahul et al., (2024) in browntop millet for number of productive tillers per plant. Ayesha et al., (2019) in foxtail millet; Priya et al., (2022) and Rahul et al., (2024) in browntop millet for grain yield per plot.
The traits like test weight (92.53%), number of productive tillers per plant (86.17%), harvest index (78.61%), grain yield per plant (78.07%), fodder yield per plant (76.77%) showed high estimates of heritability. Moderate estimates of heritability was recorded for days to 50% flowering (73.59%), plant height (69.26%), days to maturity (67.66%) and panicle length (34.93%).
Higher estimates of genetic advance as percentage of mean were recorded for number of productive tillers per plant (52.88%) followed by grain yield per plant (46.19%), harvest index (35.69%) and test weight (27.48%). Moderate estimates of GAM were recorded for plant height (15.77%) and fodder yield (18.16%).
High estimates of heritability and genetic advance as percentage of mean were recorded for test weight ( h2 : 92.53, GA: 27.48%), number of productive tillers per plant ( h2 : 86.17, GA: 52.88 %), harvest index ( h2 : 78.61, GA:35.69%) and grain yield per plant ( h2 : 78.07, GA:46.19%). High estimates of indicates the predominance of additive gene action. Therefore, phenotypic selection would be more effective for improvement of these traits. These results are in agreement with Yadav et al., (2024) in foxtail millet; Patel et al., (2018) in little millet; Rahul et al., (2024) in browntop millet for number of productive tillers per plant and grain yield per plant; Smita et al., (2016) and Patel et
Table 2. Mean, range, variability, heritability (broad sense) and genetic advance as per cent of mean of the characters
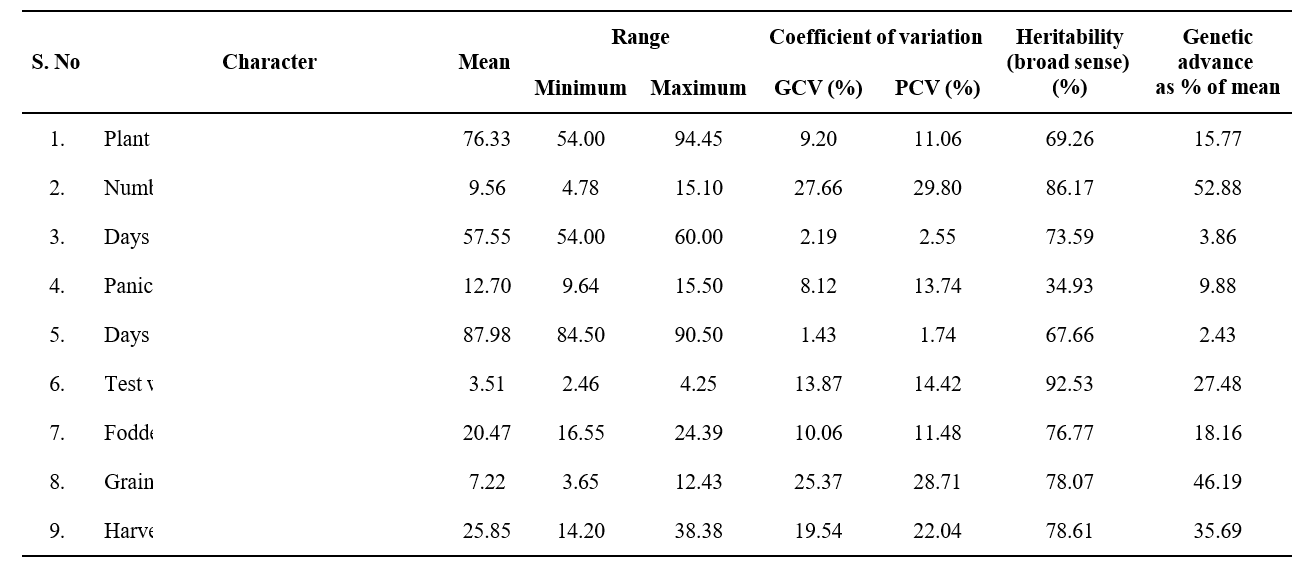
- (2018) in little millet for test weight and Katara et al. (2019) for harvest index in little millet.
The results highlight that traits viz., number of productive tillers per plant, grain yield per plant, harvest index and test weight exhibit considerable genetic variability, high heritability coupled with significant genetic advance, making them ideal traits for improvement through selection. These findings underscore the importance of integrating variability measures with heritability and genetic gain estimates to formulate effective breeding strategies.
LITERATURE CITED
Ayesha, Md., Babu, D.R., Babu, J.D.P and Rao, V.S. 2019. Studies on correlation and path analysis for grain yield and quality components in foxtail millet (Setaria italica (L.) Beauv.). International Journal of Current Microbiology and Applied Sciences. 8(4): 2173-2179.
Basappa, G.P., Muniyamma, M and Chinnappa, C.C. 1987. An investigation of chromosome numbers in the genus Brachiaria (Poaceae: Paniceae) in relation to morphology and taxonomy. Canadian Journal of Botany. 65: 2297-2309.
Katara, D., Kumar, R., Rajan, D., Chaudhari, S.B and Zapadiya, V.J. 2019. Genetic-morphological analysis in little millet (Panicum sumatrance Roth. Ex Roemer and Schult..) under different sown conditions. International Journal of Current Microbiology and Applied Sciences. 8(5): 177-189.
Patel, S.N., Patil, H.E., Modi, H.M and Singh, J.T. 2018. Genetic variability study in little millet (Panicum miliare L.) genotypes in relation to yield and quality traits. International Journal of Current Microbiology and Applied Sciences. 7(6): 2712-2725.
Priya, M.S., Madhavilatha, L. and Kumar, M.H., 2022. Genetic divergence, trait association and path analysis studies in browntop millet germplasm. Electronic Journal of Plant Breeding. 13(3): 1156-1161.
Rahul, G.K., Bhavani, P., Shyamalamma, S. and Nandini, C., 2024. Assessment of genetic diversity in germplasm collections of Browntop millet (Brachiaria ramosa (L.) Stapf.) using morphological traits. Electronic Journal of Plant Breeding. 15(1): 217-225.
Sheahan, C.M. 2014. Plant guide for browntop millet (Urochloa ramosa). USDA- Natural Resources Conservation Service, Cape May Plant Materials Center, Cape May, NJ.
Smita, S., Patil, J.V., Sunil, G and Ganapathy, K.N. 2016. Genetic variability and association analysis for grain yield and nutritional quality in foxtail millet. International Journal of Bio- Resource and Stress Management. 7 (6): 1239- 1243.
Yadav, Y., Dhoot, R., Kumhar, S.R. and Roy, C. 2024. Estimates of variability, heritability and genetic advance in foxtail millets. Annals of Arid Zone. 63(1): 121-123.
- Effect of Sowing Window on Nodulation, Yield and Post – Harvest Soil Nutrient Status Under Varied Crop Geometries in Short Duration Pigeonpea (Cajanus Cajan L.)
- Nanotechnology and Its Role in Seed Technology
- Challenges Faced by Agri Startups in Andhra Pradesh
- Constraints of Chcs as Perceived by Farmers in Kurnool District of Andhra Pradesh
- Growth, Yield Attributes and Yield of Fingermillet (Eleusine Coracana L. Gaertn.) as Influenced by Different Levels of Fertilizers and Liquid Biofertilizers
- Consumers’ Buying Behaviour Towards Organic Foods in Retail Outlets of Ananthapuramu City, Andhra Pradesh

